Abstract
The p53 tumor suppressor plays a central role in integrating cellular responses to various stresses. Tight regulation of p53 is thus essential for the maintenance of genome integrity and normal cell proliferation. Currently, several ubiquitin ligases, including the single-subunit RING-finger types—MDM2, Pirh2, and COP1—and the HECT-domain type—ARF-BP1—have been reported to target p53 for degradation. Here, we report the identification of a human Kelch domain-containing F-box protein, JFK. We showed that JFK promotes ubiquitination and degradation of p53. But unlike MDM2, Pirh2, COP1, and ARF-BP1, all of which possess an intrinsic ubiquitin ligase activity, JFK destabilizes p53 through the assembly of a Skp1-Cul1-F-box complex. Significantly, JFK inhibits p53-dependent transcription, and depletion of JFK stabilizes p53, promotes cell apoptosis, arrests cells in the G1 phase, and sensitizes cells to ionizing radiation-induced cell death. These data indicate that JFK is a critical negative regulator of p53 and represents a pathway for the maintenance of p53 levels in unstressed cells. Our experiments link the Skp1-Cul1-F-box system to p53 regulation.
Under normal cell growth conditions, the level of p53 protein is kept low through regulation of its protein stability by a number of negative regulators. Although earlier studies suggested that MDM2 is the primary factor regulating p53 turnover through mono- or poly-ubiquitination of p53, additional cellular factors have since been identified that facilitate p53 degradation through ubiquitin/proteasome-dependent mechanisms, indicating that the regulation of p53 stability is more complex than originally thought. Indeed, several other proteins, including Pirh2 (1), COP1 (2), and ARF-BP1 (3), have been reported to also promote p53 turnover. All these proteins possess an intrinsic ubiquitin ligase activity, and, interestingly, MDM2, Pirh2, and COP1 each form an autoregulatory negative feedback loop with p53.
Ubiquitination is accomplished by the ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s). E3 ubiquitin ligases include the single-subunit RING-finger type, the multi-subunit RING-finger type, and the HECT-domain type. The best characterized mammalian multi-subunit RING-finger type of ubiquitin ligase is the SCF (Skp1-Cul1-F-box) complex. Each of the SCF complexes is composed of 4 subunits: Skp1, Cul1/Cdc53, Roc1/Rbx1/Hrt1, and an F-box protein (4). Cul1 serves as a scaffold and interacts with Rbx1 at its carboxyl terminus to recruit ubiquitin-conjugated E2 (4), while the amino terminus of Cul1 interacts with Skp1, which in turn binds to an F-box protein, the subunit responsible for substrate specification (4).
Despite an expected substrate diversity and an anticipated functional divergence of the SCF complexes, thus far these ubiquitin ligases have been mainly found to function in controlling the stability of cell cycle regulators, especially those that act on the G1/S transition (5). As rapid, specific, and timely proteolysis of cell cycle regulators represents a major mechanism ensuring proper progression through the cell division cycle, the importance of the SCF ligase complexes in cell cycle control is underpinned. However, so far, the reported E3 ligases for p53 are either single-subunit RING-finger types (MDM2, Pirh2, and COP1) or HECT-domain type [ARF-BP1 (3)]; no SCF ubiquitin ligase has been reported for p53 regulation to our knowledge. Considering the key role of p53 in controlling the G1/S cell cycle checkpoint (6, 7) and the importance of the SCF complex in regulating the G1/S transition (5), the existence of such a complex for p53 is plausible.
As substrate receptors of SCF complexes, F-box proteins constitute a large family of eukaryotic proteins that feature an approximately 40-aa F-box motif (8–11). Although they may potentially influence a variety of cellular processes, F-box proteins were first described in the SCF complex (9, 12) and have since been characterized as an integral subunit of the SCF ligase complexes. Consistent with a primary function of the SCF ubiquitin ligases as regulatory machineries for the periodic proteolysis of cell cycle regulators, several F-box proteins, including Skp2 (13–17), β-TrCP (5, 18), and Fbw7 (19–24), have been shown to target cell cycle regulators.
A total of 68 F-box proteins have been found in humans (8), but the function of most of them remains unknown. The F-box proteins fall into 3 major classes: Fbws containing WD40 repeats, Fbls containing leucine-rich repeats, and Fbxos containing other types of domains, such as the CHORD domain, the TPR-like domain, the CASH domain, the SPRY domain, and Kelch repeats (8, 10). Although Kelch domain-containing F-box proteins make up the majority of the F-box proteins in Arabidopsis species (25), this type of F-box is rare in mammals. Here we report on the identification and characterization a Kelch domain-containing F-box protein, JFK, in humans. We show that JFK promotes p53 turnover through assembly of an SCF complex, revealing a pathway for the control of p53 degradation and providing a link between the SCF complex and p53 regulation.
Results and Discussion
Cloning and Characterization of JFK.
During the process of yeast 2-hybrid screening of a mammary cDNA library (Clontech) for steroid receptor co-activator-interacting protein, we cloned a gene in our laboratory. This gene is mapped to chromosome 1p16.23-p16.11. Its cDNA is 3,011 bp in length (GenBank accession no. BC043410) and contains an ORF encoding for a protein of 717 aa. The predicted molecular weight of the protein is approximately 77 kDa, with a theoretical isoelectric point 7.07. Bioinformatics analyses indicated that this protein contains an F-box motif in its N terminus and a Kelch domain downstream of the F-box [supporting information (SI) Fig. S1A]. We named this gene JFK because the human genome encodes apparently Just one F-box and Kelch domain-containing protein.
To confirm the existence of JFK transcript(s) and to examine the tissue distribution of JFK, we first analyzed the expression of JFK mRNA by Northern blotting using Clontech multiple tissue blots. This analysis indicated that JFK transcribes a message that is approximately 3.0 kb in length in various human tissues and at different levels of enrichment (Fig. S1B). Specifically, the level of JFK mRNA expression was highest in heart and skeletal muscle, moderate in placenta, liver, and pancreas, and low in brain, lung, and kidney. To confirm the predicted molecular weight of JFK protein, FLAG-tagged JFK expression vector (FLAG-JFK) was transfected into human osteosarcoma U2OS cells. Analyzing cellular extracts by Western blotting with an anti-FLAG antibody indicated that the cloned cDNA encodes for a protein with an apparent molecular weight of approximately 77 kDa (Fig. S1C Left). Western blotting analysis of the cellular extracts of vector-transfected U2OS cells with polyclonal antibodies against JFK that we generated in rabbit by using a C-terminal epitope of JFK protein (CYPKTNALYFVRAKR) showed that the endogenous JFK is a protein of approximately 77 kDa (Fig. S1C Right), confirming its predicted molecular weight.
JFK Is Associated with Skp1, Cul1, and Rbx1, the Components of the SCF Complex.
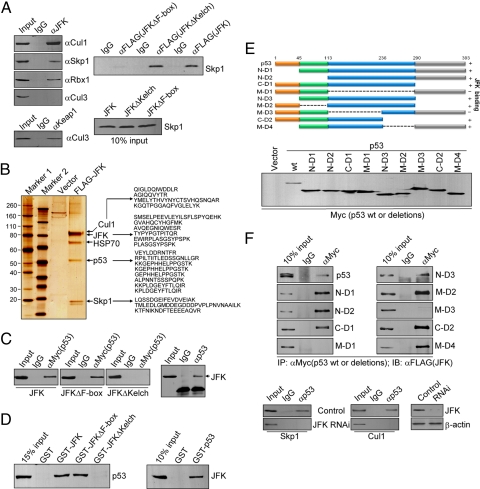
As stated before, F-box proteins were first found in SCF complexes and have since been found to be an integral component of a number of SCF ubiquitin ligase complexes as a substrate receptor. One structural feature in particular, the presence of an F-box motif in JFK, prompted us to investigate whether JFK also functions in the context of an SCF complex. For this purpose, U2OS cellular lysates were immunoprecipitated with anti-JFK followed by immunoblotting with antibodies against Cul1, Skp1, or Rbx1. The results indicated that JFK was indeed co-immunoprecipitated with Cul1, Skp1, and Rbx1 but not with Cul3, although Keap1, an adaptor for Cul3-based E3 ligase (26), was efficiently co-immunoprecipitated with Cul3 (Fig. 1A Left). This suggests that JFK is specifically associated with Skp1, Cul1, and Rbx1, the constituents of the SCF complex. In addition, the association of JFK with the components of the SCF complex is dependent on its F-box, as an F-box-deleted JFK mutant (JFKΔF-box) failed to be co-immunoprecipitated with Skp1, whereas a Kelch domain-deleted JFK mutant (JFKΔKelch) still retained the capacity to be co-immunoprecipitated with Skp1 (Fig. 1A Right). This is consistent with the feature of all F-box proteins that are found in SCF complexes, in which the F-box protein interacts with Skp1 through the F-box motif (4, 12). Collectively, these experiments support the hypothesis that JFK is associated with Skp1, Cul1, and Rbx1 and may thus function via assembly of an SCF complex.
Fig. 1.
JFK is associated with the SCF complex and interacts with p53. (A) (Left) JFK is associated with Skp1, Cul1, and Rbx1 in vivo. U2OS cellular extracts were immunoprecipitated with anti-JFK or anti-Keap1 followed by immunoblotting (IB) with anti-Skp1, -Cul1, -Rbx1, or -Cul3. (Right) F-box of JFK is required for its interaction with Skp1. U2OS cells were transfected with FLAG-JFK, FLAG-JFKΔF-box, or FLAG-JFKΔKelch. Cellular extracts were prepared and immunoprecipitation was performed with anti-FLAG followed by immunoblotting with anti-Skp1 (Upper). Ten percent of input is shown (Lower). (B) JFK is associated with p53 in vivo. Lysates from U2OS cells stably expressing FLAG(3x)-JFK were immunoprecipitated with anti-FLAG. Bound proteins were eluted with FLAG peptide, resolved on NuPAGE 4%–12% Bis-Tris gels were silver stained and analyzed by mass spectrometry. The identified peptides are listed. (C) The Kelch domain of JFK is required for its interaction with p53. H1299 cells were co-transfected with Myc-p53 and FLAG-JFK, FLAG-JFKΔKelch, or FLAG-JFKΔF-box. Cellular lysates were immunoprecipitated with anti-Myc followed by immunoblotting with anti-FLAG (Left). Cellular extracts were also prepared from untransfected U2OS cells for co-immunoprecipitation assays with anti-p53 followed by immunoblotting with anti-JFK (Right). (D) JFK interacts with p53 in vitro. GST pull-down experiments were performed with GST-fused JFK or JFK mutants and in vitro transcribed/translated p53 (Left) or with GST-p53 and in vitro transcribed/translated JFK (Right). (E) Mapping the domain of p53 that is required for its interaction with JFK. H1299 cells were co-transfected with FLAG-JFK and Myc-tagged WT p53 or p53 deletion mutants. Cellular extracts were immunoprecipitated with anti-Myc followed by immunoblotting with anti-FLAG. The schematic diagrams of p53 truncations and 10% inputs of these truncations are shown. (F) The interaction between JFK and p53 is independent of the interaction between JFK and Cul1/Skp1. U2OS cellular lysates with or without JFK siRNA treatment were immunoprecipitated with anti-p53 followed by immunoblotting with antibodies against the indicated proteins.
JFK Physically Interacts with p53.
To gain further insight into the biological function of JFK, lysates from U2OS cells stably expressing FLAG(3x)-JFK were immunoprecipitated with anti-FLAG, and bound proteins were eluted with FLAG peptide and resolved by SDS/PAGE (27–30). Mass spectrometric analysis identified the presence of Skp1 and Cul1 in the JFK-containing protein complex (Fig. 1B), confirming the association of JFK with the SCF complex in vivo. Interestingly, 19 matching peptides from the tumor-suppressor protein p53 were also identified in the JFK-containing protein complex (SI Materials and Methods), suggesting that JFK is associated with p53 in vivo.
To confirm an in vivo association between JFK and p53, p53-null H1299 cells were co-transfected with Myc-tagged p53 (Myc-p53) and FLAG-tagged JFK or JFK mutants. Cellular lysates were immunoprecipitated with anti-Myc followed by immunoblotting with anti-FLAG. The results indicated that p53 was co-immunoprecipitated with JFK and JFKΔF-box but not with JFKΔKelch (Fig. 1C Left), supporting the conclusion that JFK and p53 physically interact and indicating that the Kelch domain of JFK is required for this interaction. In addition, an interaction between endogenous JFK and p53 was also detected in U2OS cells by immunoprecipitation with anti-p53 followed by immunoblotting with anti-JFK (Fig. 1C Right). Moreover, the interaction between JFK and p53 is direct, as GST pull-down experiments (31, 32) revealed that in vitro transcribed/translated p53 strongly binds to GST-JFK and, reciprocally, in vitro transcribed/translated JFK strongly binds to GST-p53 (Fig. 1D). In support of the observation that the Kelch domain of JFK is required for the interaction of JFK with p53, GST pull-down assays showed that JFKΔKelch failed to pull down p53 whereas JFKΔF-box was able to do so (Fig. 1D). Taken together, these data indicate that p53 and JFK interact in vivo and in vitro.
To investigate the molecular detail involved in the interaction of p53 with JFK, H1299 cells were co-transfected with FLAG-JFK and Myc-p53 or Myc-tagged serial deletion mutants of p53. Cellular lysates were immunoprecipitated with anti-Myc, followed by immunoblotting with anti-FLAG. These experiments demonstrated that neither the N-terminal fragment nor the C-terminal fragment of p53 was capable of binding JFK, whereas the central core region extending from residues 113 to 236 exhibited JFK binding capacity (Fig. 1E). These results suggest that this latter region is responsible for p53 binding to JFK.
Finally, to investigate whether the interaction between p53 and Cul1/Skp1 is dependent on that of JFK and p53, U2OS cellular lysates with or without JFK siRNA treatment were immunoprecipitated with anti-p53 followed by immunoblotting with antibodies against Cul1 or Skp1. The data indicated that p53 was not co-immunoprecipitated with Cul1 and Skp1 in the absence of JFK, supporting the argument that the interaction between p53 and Cul1/Skp1 depends on that between JFK and p53 (Fig. 1F).
JFK Promotes p53 Protein Degradation Through an SCF-Dependent Pathway.
As mentioned earlier, F-box proteins are an integral component of the SCF ligase complexes that mainly function in the periodic proteolysis of cell cycle regulators. To determine the functional significance of the physical interaction between JFK and p53, we examined the effect of JFK on the steady-state level of p53 protein. In addition to p53, we also measured other cell cycle regulators including c-Myc, c-Jun, NF-κB, cyclin D1, cyclin D2, cyclin D3, cyclin E, and p27. Western blotting analysis of the cellular lysates of U2OS cells transfected with FLAG-JFK with antibodies against these proteins revealed that, whereas the expression of the rest of the proteins was essentially unchanged under JFK overexpression, the level of p53 was significantly reduced (Fig. 2A). Moreover, the reduction in p53 protein level associated with JFK overexpression was probably through a proteasome-mediated protein degradation mechanism, as the effect could be effectively blocked by a proteasome specific inhibitor, MG132. As controls, there were no evident changes in p53 protein steady-state levels under overexpression of Fbw7 (the F-box protein for cyclin E, c-Myc, c-Jun, Notch-1 and -4), β-TrCP (the F-box protein for Emi1, Cdc25a, Wee1, β-catenin, and IκB-family members), or Skp2 (the F-box protein for p27, p21, p57, and p130; Fig. 2B Upper). In addition, the decreased level of p53 was observed only in cells transfected with WT JFK but not JFKΔF-box or JFKΔKelch (Fig. 2B Lower). Together with the observations that JFK interacts with p53 in vitro and in vivo, these data indicate that p53 is a target and a substrate for JFK.
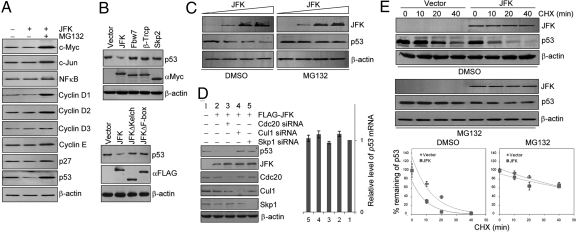
Fig. 2.
JFK promotes p53 turnover via an SCF-dependent mechanism. (A) JFK destabilizes p53 protein. U2OS cells were transfected with JFK. Cellular extracts were prepared under no treatment or treatment with MG132 and analyzed by Western blotting for protein expression with antibodies against the indicated proteins. (B) (Upper) p53 degradation is specific to JFK. U2OS cells were transfected with Myc-tagged JFK, Fbw7, β-TrCP, or Skp2. Cellular extracts were prepared and Western blotting was performed with anti-p53. (Lower) WT JFK but not JFK mutants promote p53 degradation. U2OS cells were transfected with FLAG-JFK, FLAG-JFKΔKelch, or FLAG-JFKΔF-box. Cellular extracts were prepared and Western blotting was performed with anti-p53. (C) JFK decreases the steady-state level of exogenous p53 protein. H1299 cells were co-transfected with a constant amount of p53 and increasing amounts of FLAG-JFK. Forty-eight hours after transfection, the cells were treated with vehicle (DMSO) or MG132 for 6 h before cells were collected for Western blotting analysis. (D) (Left) The destruction of p53 by JFK is mediated by an SCF complex. U2OS cells were transfected with JFK together with siRNAs for knocking down Cul1, Skp1, or Cdc20 as indicated. Cellular proteins were extracted for Western blotting analysis with antibodies against the indicated proteins. (Right) JFK does not alter p53 mRNA levels. Total RNAs were extracted from the above-described U2OS cells and analyzed for p53 expression by real-time RT-PCR. The data were normalized to GAPDH. Each bar represents the mean ± SD for triplicate experiments. (E) JFK decreases p53 half-life. U2OS cells were transfected with vector or FLAG-JFK. Forty-eight hours after transfection, cells were treated with vehicle (DMSO) or MG132 for 6 h before addition of cycloheximide (CHX; 50 μg/mL). Cells were then harvested for Western blotting analysis at the designated times after CHX treatment. Quantitation was done by densitometry and expressed as signals of p53/β-actin. Each point represents the mean ± SD for triplicate experiments.
To further support this deduction, the potential of JFK to modulate the steady-state levels of p53 protein was assessed in H1299 cells by expressing a constant amount of p53 and increasing amounts of JFK. The results show that increased levels of JFK protein were associated with decreased levels of p53, but only in the absence of MG132; in the presence of MG132, increased levels of JFK had little effect on the levels of p53 (Fig. 2C). In addition, whereas knockdown of Cdc20, the activator subunit of the mitotic APC (anaphase-promoting complex), had no effect on JFK-associated p53 degradation in U2OS cells, knockdown of Cul1 or Skp1 led to a diminished effect on JFK-associated p53 degradation in these cells (Fig. 2D Left). The decreased p53 protein expression under JFK overexpression was not a result of p53 mRNA degradation, as real-time quantitative RT PCR measurements indicated that JFK overexpression did not result in a decreased p53 mRNA level in U2OS cells (Fig. 2D Right). Furthermore, cycloheximide chase assays in U2OS cells revealed that JFK overexpression was associated with a decreased p53 half-life in the absence but not in the presence of MG132 (Fig. 2E). Together with the observation that JFK is associated with the components of the SCF complex, these experiments support the hypothesis that JFK-promoted destruction of p53 operates through an ubiquitin-proteasome pathway and via an SCF-dependent mechanism.
JFK Promotes p53 Ubiquitination.
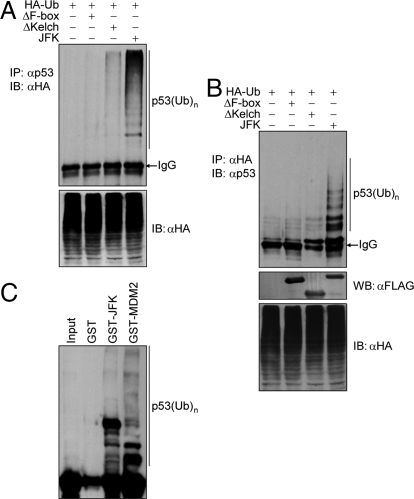
To further substantiate a receptor-substrate relationship and to support the ubiquitin-dependent mechanism of p53 degradation by JFK, we next determined whether JFK-mediated p53 degradation is a consequence of p53 ubiquitination. To this end, U2OS cells were co-transfected with FLAG-JFK or JFK mutants together with HA-tagged ubiquitin. Immunoprecipitation of the cellular lysates with anti-p53 and immunoblotting with anti-HA detected increased levels of ubiquitinated p53 species under expression of WT JFK but not that of JFKΔF-box or JFKΔKelch (Fig. 3A). Reciprocal immunoprecipitation in U2OS cells with anti-HA and immunoblotting with anti-p53 detected strong p53 ubiquitination under JFK overexpression (Fig. 3B). These data indicate that JFK promotes p53 ubiquitination in vivo. In vitro ubiquitination assays with bacterially expressed GST-JFK and in vitro transcribed/translated p53 revealed that JFK protein is capable of ubiquitinating p53 in vitro (Fig. 3C). Together, these data strongly support the hypothesis that JFK targets p53 for degradation involving p53 ubiquitination. MDM2 was included in these experiments as a positive control.
Fig. 3.
JFK promotes p53 ubiquitination. (A) U2OS cells were co-transfected with the indicated plasmids. Forty-eight hours after transfection, cells were treated with MG132 for 6 h before cellular extracts were prepared for co-immunoprecipitation assays with anti-p53 followed by immunoblotting with anti-HA. (B) U2OS cells were co-transfected with the indicated plasmids. Forty-eight hours after transfection, cells were treated with MG132 for 6 h before cellular extracts were prepared for co-immunoprecipitation assays with anti-HA followed by immunoblotting with anti-p53. The expression of JFK, JFKΔKelch, and JFKΔF-box was examined by Western blotting (WB). (C) JFK ubiquitinates p53 in vitro. Bacterially expressed GST-JFK or GST-MDM2 was incubated with in vitro transcribed/translated p53 for ubiquitination assays. The reaction mixture was then resolved on SDS/PAGE and exposed.
Biological Significance of p53 Destabilization by JFK.
To investigate the biological consequences of JFK-promoted p53 degradation, we first assessed the effect of JFK on the trans-activation activity of p53. In these experiments, H1299 cells with forced expression of p53 were transfected with a luciferase gene driven by a synthetic promoter containing multiple p53 binding sites (pG13-Luc) (33) together with WT JFK or JFK mutants. Luciferase reporter assays revealed that JFK negatively regulated p53 trans-activation activity, and the F-box and Kelch domains of JFK were essential in this regulation (Fig. 4A). We then examined the effect of JFK on the transcription of natural p53 target genes. U2OS cells stably expressing JFK were subjected to real-time RT-PCR analysis of the expression of p21 and Bax. The results indicated that JFK overexpression was associated with decreased transcriptions of p21 and Bax. Consistent with this, the level of p53 protein was decreased and p21 and Bax protein expression were also reduced in these cells (Fig. 4B). To further explore the functional link between the E3 ligase activity of JFK and the trans-activation activity of p53, loss-of-function experiments were performed to examine the effect of JFK on the transcription regulation of p21 expression by p53 via knockdown of JFK as well as the other p53 E3 ligases, including MDM2, Pirh2, COP1, and ARF-BP1, either individually or combinatorially. For these experiments, 3 to 4 siRNAs for each of the E3 ligases were designed and were introduced into U2OS cells via transfection. The mRNA expression of these E3 ligases was then measured by quantitative real-time RT-PCR. The most effective siRNAs were chosen and the experiments were optimized to achieve a similar knockdown efficiency (≈80%) on the level of mRNA for all these E3 ligases (Fig. S2A Left). The knockdown efficiency was also examined by Western blotting analysis of the protein expression of all of the E3 ligases except for ARF-BP1 (Fig. S2A Right), for which no commercial antibody is available. We measured the mRNA and protein expression of the p53 target gene, p21, Bax, and PUMA by real-time RT-PCR and Western blotting, respectively, in U2OS cells under knockdown of the E3 ligases. The results of these experiments indicated that JFK knockdown led to an increase in p21 transcriptional activity threefold relative to the control (Fig. 4C Upper). In comparison, knockdown of Pirh2 or COP1 was associated with increases in p21 transcriptional activity by approximately 1.5 to 2 fold, and ARF-BP1 knockdown led to a threefold increase, whereas MDM2 knockdown resulted in a fourfold increase, which may be a reflection of the ability of MDM2 not only to increase p53 turnover but also to repress the trans-activation activity of p53 (1, 2). Double knockdown of JFK and Pirh2, JFK and COP1, or JFK and ARF-BP1 resulted in further elevations in p21 transcriptional activity by four to seven fold. However, simultaneous knockdown of JFK and MDM2 yielded an eightfold increase in p21 transcriptional activity. This was not further enhanced by additional ARF-BP-1 knockdown, suggesting that p21 transcriptional activity had reached a maximum in this particular system whereas JFK, COP1, and Pirh2 co-knockdown resulted in a lower increase relative to former combination. The effect of the E3 ligase knockdown on the mRNA expression of Bax and PUMA was similar to that of p21. Consistent with the increased p53 target genes transcriptional activity, the levels of p53, p21, and Bax proteins were elevated in these systems, as measured by Western blotting (Fig. 4C Lower). Furthermore, individually knocking down JFK, MDM2, Pirh2, COP1, or ARF-BP1 resulted in increases in the steady-state levels of p53 and p21 in normal mammary epithelial MCF-10A cells, indicating that each ligase may exert an independent activity in the negative regulation of p53 in normal cells (Fig. S2B). Collectively, these data suggest that the effect of p53 destabilization by JFK could extend to p53 transcriptional activation activity and may thus instigate physiological responses.
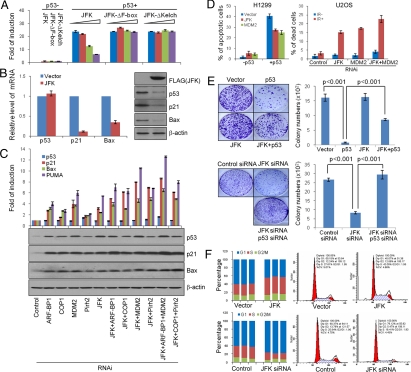
Fig. 4.
Biological significance of p53 regulation by JFK. (A) The effect of JFK on p53 trans-activation activity. H1299 cells were co-transfected with p53 and WT JFK or JFK mutants together with pG13-Luc. Cellular lysates were prepared and luciferase activity was measured. Each bar represents the mean ± SD for triplicate experiments. (B) The effect of JFK on p21 and Bax expression. The mRNA or protein level of p21 and Bax were analyzed by real-time RT-PCR or Western blotting, respectively, in U2OS cells stably expressing JFK. Each bar represents the mean ± SD for triplicate experiments. (C) JFK knockdown stabilizes p53 and activates the transcription of p21, Bax, and PUMA. U2OS cells were transfected with siRNA oligonucleotides for JFK and/or for MDM2, Pirh2, COP1, or ARF-BP1 as indicated. Total RNAs were prepared for real time RT PCR analysis of mRNA expression of p53, p21, Bax, and PUMA (Upper). The data were normalized to GAPDH. Each bar represents the mean ± SD for triplicate experiments. Cellular lysates were prepared for Western blotting analysis with antibodies against the indicated proteins (Lower). (D) (Left) JFK overexpression suppresses p53-induced cell apoptosis. H1299 cells with forced expression of p53 were transfected with JFK or MDM2 before double staining with annexin V and propidium iodide. Cell apoptosis was determined by flow cytometry. (Right) JFK and/or MDM2 knockdown sensitizes U2OS cells to IR-induced cell death. U2OS cells were pretreated with the indicated siRNA before 20 Gy IR. Cell death was determined by propidium iodide staining. Each bar represents the mean ± SD for triplicate experiments. (E) The effect of JFK on the growth-suppressing activity of p53. H1299 (Upper) or U2OS (Lower) cells stably expressing JFK were transfected with p53 or pretreated with the indicated siRNA oligonucleotides. Cells were maintained in G418-containing medium for 14 d before staining with crystal violet and counting for colony numbers. Each bar represents the mean ± SD for triplicate experiments. Statistical analysis was performed with one-tailed unpaired t test. (F) The effect of JFK on the cell cycle profile of U2OS cells. U2OS cells stably expressing JFK or transfected with JFK siRNA were subjected to cell cycle analysis by flow cytometry. The data represent 3 independent experiments. The representative cytometric results from these experiments are shown.
Next, we examined the effect of overexpression of JFK or MDM2 on p53-induced apoptosis. For this purpose, H1299 cells ectopically expressing p53 were transfected with JFK or MDM2. The cells were then double-stained with annexin V and propidium iodide and subjected to flow cytometric analysis. The results revealed that the forced expression of p53 caused approximately 40% of H1299 cells to undergo apoptosis (Fig. 4D Left). However, overexpression of JFK or MDM2 resulted in a decrease in apoptotic cells by approximately 13% or approximately 16%, respectively. We then tested whether knockdown of JFK and/or MDM2 would restore a normal DNA-damage response in U2OS cells, as these cells are known to be highly resistant to ionizing radiation (IR)-induced cell death (34). In these experiments, U2OS cells were transfected with siRNAs to target JFK and/or MDM2, treated with 20 Gy IR, and subsequently used for cell death assays by propidium iodide staining. As expected, U2OS cells pretreated with control siRNA were highly resistant to IR-induced cell death; however, cells pretreated with JFK siRNA or MDM2 siRNA resulted in a significant increase in cell population that were sensitive to IR-induced cell death, and simultaneous depletion of JFK and MDM2 resulted in a greater sensitization to cell death by IR (Fig. 4D Right).
We then investigated the effect of JFK on p53-dependent growth suppression. For this purpose, H1299 cells stably expressing JFK were transfected with a p53 expression construct that also carries the gene for neomycin resistance. Cells were maintained in culture medium supplemented with 1 mg/mL G418 for 14 d and stained with crystal violet for colony counting. Colony formation assays revealed that the expression of p53 in H1299 cells resulted in a significant decrease in colony number compared with the parental H1299 cells, which do not express p53 (Fig. 4E Upper). However, overexpression of JFK in p53-tranfected H1299 cells resulted in a significant increase in colony number. Conversely, knockdown of the expression of JFK in U2OS cells resulted in a significant decrease in colony number, which could be rescued by simultaneous knockdown of p53 (Fig. 4E Lower). These data suggest that the E3 ligase activity of JFK is required to block the growth suppressor function of p53.
Given that overexpressing JFK reduced the amount of endogenous p53 protein and that RNAi-mediated knockdown of JFK increased the amount of endogenous p53 protein, we wished to determine if JFK could affect the cell cycle. U2OS cells stably expressing JFK or with JFK knockdown were used. The effect of the gain of function and loss of function of JFK on the cell cycle profile was analyzed by propidium iodide staining and flow cytometry. Compared with control, JFK overexpression was associated with a decreased cell population in G1 and an increased cell population in the S phase (Fig. 4F). This was reflected by a decrease in the G1/S ratio from 2.28 to 1.03. Conversely, U2OS cells exposed to JFK siRNA exhibited an increase in the proportion of cells in G1 and a decrease in the proportion of cells in the S phase (G1/S, from 2.12 to 4.95). These data are consistent with the property of arresting cells in the G1 phase of the cell cycle by p53. Taken together, these experiments support roles of JFK in the regulation of p53-mediated transcriptional activation, p53-mediated cell apoptosis/death, growth suppression, and checkpoint control.
It is interesting to note that JFK protein expression is higher in cancer cell lines compared with that in normal cell lines (Fig. S1D). The potential role of JFK and its functional connection with p53 in carcinogenesis need future investigations. In addition, it appears that the regulation of p53 by JFK is not cell cycle-dependent (Fig. S3). Future studies are warranted to investigate the relationship among the various p53 E3 ligases and their functional specificity.
Materials and Methods
A ubiquitin-protein conjugation kit (Boston Biochem) was used for the in vitro ubiquitination assay. Briefly, 5 μL of rabbit reticulocyte lysate-translated p53 was incubated in the absence or presence of bacterially expressed GST-JFK, GST-MDM2, or GST (3 μg), 8 μg of fraction I, 8 μg of fraction II, 26 μg of ubiquitin, 4 μM ubiquitin aldehyde, and 2.5 μL of energy solution (10×) in a 25-μL volume. After incubation at 37 °C for 30 min, the reactions were stopped with 25 μL of 2× sample loading buffer and resolved on 8% SDS/PAGE. Detailed descriptions of materials and methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
This work was supported by National Natural Science Foundation of China Grants 30830032, 30621002, and 30470912 (to Y.S.) and Ministry of Science and Technology of China 863 Program Grant 2006AA02Z466 and 973 Program Grants 2005CB522404 and 2007CB914503 (to Y.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901864106/DCSupplemental.
References
- 1.Leng RP, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 2.Dornan D, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 3.Chen D, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 4.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 5.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–333. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 6.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 7.Kastan MB, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 8.Jin J, et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai C, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 10.Cenciarelli C, et al. Identification of a family of human F-box proteins. Curr Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 11.Craig KL, Tyers M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol. 1999;72:299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 12.Skowyra D, et al. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 13.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 14.Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsvetkov LM, et al. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- 16.Yam CH, et al. Regulation of cyclin A-Cdk2 by SCF component Skp1 and F-box protein Skp2. Mol Cell Biol. 1999;19:635–645. doi: 10.1128/mcb.19.1.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kossatz U, et al. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev. 2004;18:2602–2607. doi: 10.1101/gad.321004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin J, et al. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koepp DM, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 20.Welcker M, et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell. 2003;12:381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 21.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 22.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yada M, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 25.Lechner E, et al. F-box proteins everywhere. Curr Opin Plant Biol. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, et al. Differential gene regulation by the SRC family of coactivators. Genes Dev. 2004;18:1753–1765. doi: 10.1101/gad.1194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H, et al. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature. 2005;438:981–987. doi: 10.1038/nature04225. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, et al. The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J. 2006;25:4223–4233. doi: 10.1038/sj.emboj.7601306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, et al. SIP, a novel ankyrin repeat containing protein, sequesters steroid receptor coactivators in the cytoplasm. EMBO J. 2007;26:2645–2657. doi: 10.1038/sj.emboj.7601710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi B, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27:5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin N, et al. Molecular mechanisms involved in the growth stimulation of breast cancer cells by leptin. Cancer Res. 2004;64:5870–5875. doi: 10.1158/0008-5472.CAN-04-0655. [DOI] [PubMed] [Google Scholar]
- 33.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 34.Allan LA, Fried M. p53-dependent apoptosis or growth arrest induced by different forms of radiation in U2OS cells: p21WAF1/CIP1 repression in UV induced apoptosis. Oncogene. 1999;18:5403–5412. doi: 10.1038/sj.onc.1202931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.