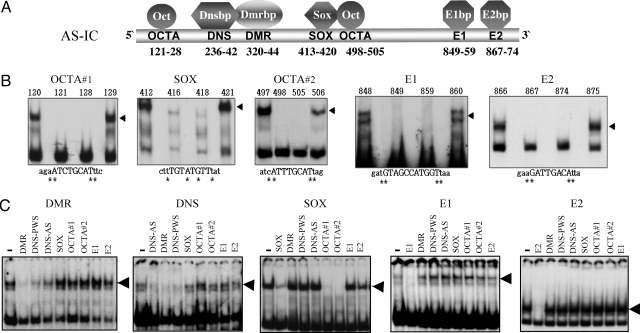
Fig. 1.
Cis elements and protein binding factors in AS-IC. (A) Names and positions of the cis elements and binding proteins in AS-IC (position 1 corresponds to nucleotide 22716614 of chr. 15, according to hg18 at http://genome.ucsc.edu/cgi-bin/hgGateway). (B) Representative band-shift assays used to characterize the binding specificity of each element. All assays were performed with nuclear extracts of mouse ES cells. Numbers above the lanes indicate the position of the mutation in the AS-IC sequence. Mutations were single base replacements G↔ T and C↔ A. The mutated bases are marked by an asterisk. Capital letters indicate bases that are part of the binding sequence. Lowercase letters represent the sequence flanking the element. The shifted bands are marked by arrowheads. Other mutated oligomers, as well as no-extract controls, were also used. (C) Competition band-shift experiments were used to determine binding cooperation between different cis elements. Labeled oligomers for each experiment are listed above each picture. The left lane of each gel represents a no-competition control. Lanes to the right represent the effect of various unlabeled competitors (vertical lettering) that were included in 100× molar excess. Loss of bands (arrowheads) indicates competition. DNS-AS and DNS-PWS represent the DNS elements on AS-IC and PWS-IC, respectively. A reciprocal experiment for OCTA competition was also performed, with results similar to that of SOX.