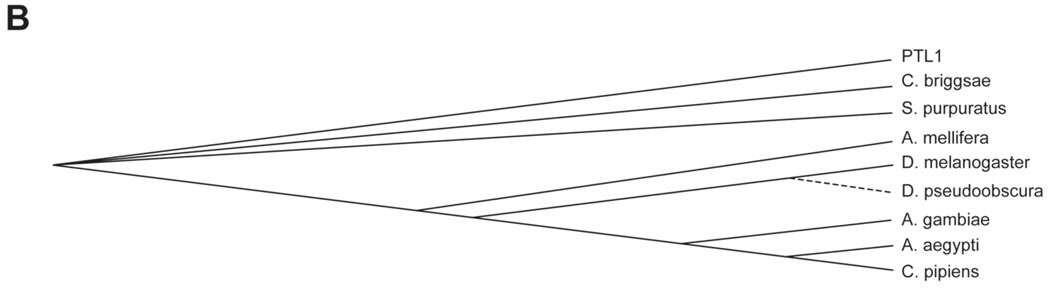
Figure 1.
Comparisons of invertebrate tau-like proteins. A) Protein sequence alignments of invertebrate tau-like proteins using Accelyrs Discovery Studio (v. 1.7) and a BLASTp protocol followed by a Clustal W progressive pairwise alignment algorithm. Dark blue indicates identical amino acid residues; lighter blue indicates similar amino acid residues (conservative substitutions). Dashed boxed regions indicate putative microtubule binding regions showing strong similarities among the sequences. Solid line box indicates the fifth putative microtubule binding region found in genus Caenorhabditis. B) Sequence similarity tree based on protein sequences compared and aligned using Accelyrs Discovery Studio multiple sequence alignment protocol followed by an unrooted neighbor joining method (Saitou and Nei, 1987) with bootstrapping (n=1000 iterations). Organisms compared (with species/protein ID): Drosophila melanogaster (NP651575), Drosophila pseudoobscura (XP001357893), Aedes aegypti (XP001655927), Culex pipiens quinquefasciatus (XP001863669), Anopheles gambiae (XP321540), Apis mellifera (XP393045), Strongylocentrotus purpuratus (XP788973), C. elegans (PTL1) and Caenorhabditis briggsae (CAE69200.1).