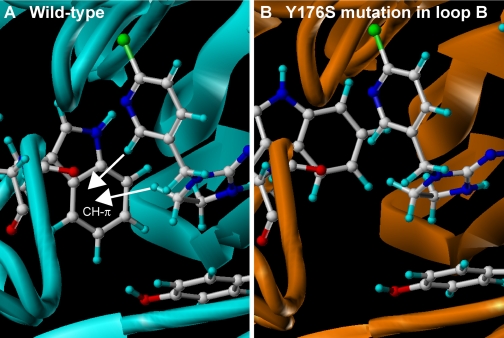
Fig. 7.
LBD homology models of wild-type α2β1 nicotinic acetylcholine receptor from the peach potato aphid M. persicae (A) and Y176S mutant (B). Models were constructed according to Toshima et al. (2009). Modeling of the N-terminal region of M. persicae α2β1 and its Y176S mutant nAChRs was carried out using the molecular modeling software package Sybyl (version 7.3; Tripos Associates, Inc.) and the homology modeling software PDFAMS Ligands & Complex (version 2.1; In-Silico Sciences, Inc., Tokyo, Japan) in the ligand and complex mode. Both α2 and β1 subunits were aligned with the Ls-AChBP bound by imidacloprid (PDB code 2ZJU). In the second step, the three-dimensional structures of the wild-type and the Y176S mutant LBD-imidacloprid complexes were generated based on the sequence alignment and the coordinates of the AChBP and imidacloprid using the simulated annealing method (Kirkpatrick et al., 1983). The coordinates of imidacloprid were fixed during the simulated annealing. The receptor model constructed in this way was energy-minimized for 5000 iterations of conjugated gradients using the force field and partial charges of the molecular mechanics MMFF94 (Halgren 1999a,b) using Sybyl. Residues within a 10-Å radius of the centrally located imidacloprid, as well as imidacloprid itself, were treated as flexible entities except the C=N-NO2 moiety, which was fixed during energy minimization. In addition, residues within 10 to 15 Å radius of the ligand were considered rigid entities to speed up the computation. Other residues were ignored in energy minimization. Carbon, hydrogen, nitrogen, oxygen, and chlorine atoms of amino acids and imidacloprid are colored white, light blue, blue, red, and blue-green, respectively. Note that the CH-π hydrogen bonds with the tryptophan residue in loop B are reduced by this mutation, consistent with enhanced imidacloprid-resistance in pests.