Fig. 9.
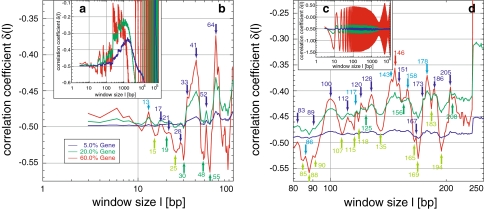
Appearance and simulation of the nucleosomal fine-structure of genomes: the fine-structure present in all human sequences (Fig. 1) is in agreement with the pattern found in simulations using a consensus nucleosomal binding sequence (a, b, d) organized in a block/gene fashion (Fig. 7). The positions of the local maxima are mostly the same as in the human genome (dark numbers/arrows are in agreement within ±1 bp), whereas the similarity of the position of the local minima is difficult to compare as they smear out in the human sequence due to the block structure of genomes (Fig. 1). Use of a mixture of two special sequence motifs results in highly ordered periodicities of 10 bp, attributable to the helical pitch and the base pairs bound to the nucleosomal core (c). The appearance, visibility, as well as the degree of correlation is again proportional to the concentration of the blocks/genes in the random sequence (see legend in b), leading also to a general maximum and oscillations of δ(l) (a, embedding hull in c)