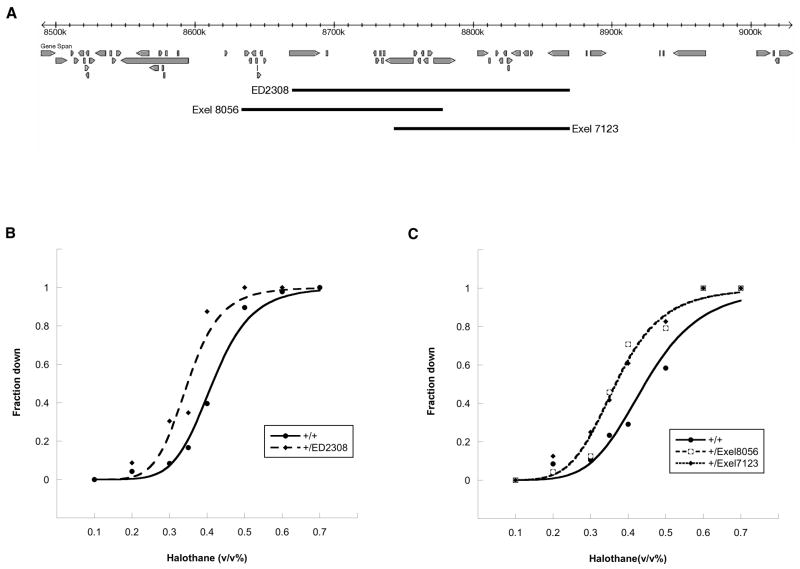
Fig. 5.
Hypersensitivity to halothane associated with multiple deletions involving one region of the genome. (A) An overview of a 540 kb segment of chromosome arm 2R. Beneath the scale ruler is shown the rough position of genes predicted= to lie in this region, the extent of the DrosDel ED2308 deletion described in Table 1, and the extent of two independently derived deletions from the Exelixis collection.(B, C) Concentration-response curves. The tested lines were generated by crossing the Canton-S wild-type strain to the indicated deletion strain or, in the case of the control heterozygotes, to the parental line from which the deletion was derived. Halothane sensitivity was assessed as described in Materials and Methods. Compared to the corresponding control line (+/+), there is a leftward shift in the EC50 value of the DrosDel deletion heterozygote (B) and both of the heterozygotes made with deletions from the Exelixis collection (C). The magnitude of this shift for +/ED2308, +/Exel8056, and +/Exel7123 is 16.6%, 17.7%, and 17.3%, respectively.