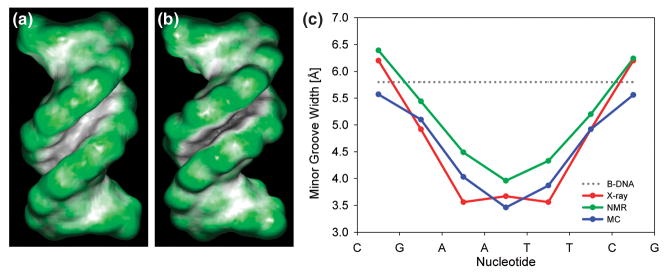
Figure 2. Sequence-dependence of minor groove shape for Dickerson dodecamer.
– Ideal B-DNA (A) and the Dickerson dodecamer, PDB code 1duf (B), differ in minor groove shape as shown by the color coding of the molecular surface (green for convex, black/grey for concave surfaces). (C) The intrinsically narrow minor groove in the center of the Dickerson dodecamer is predicted by MC simulations using ideal B-DNA as a starting conformation. The X-ray data represent an average of the 15 available crystal structures, symmetrized based on the palindromic sequence CGCGAATTCGCG in order to remove crystal packing effects. The NMR data represent an average of 10 structures that included dipolar coupling data in their structure determination. Minor groove width is calculated with the CURVES program [55].