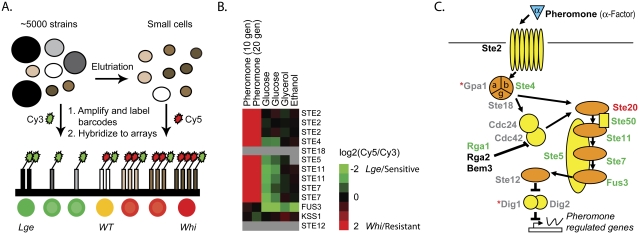
Figure 4.
The pheromone pathway affects cell size during exponential growth. (A) Mixed yeast haploid deletion pools were elutriated to enrich for small cells. Unique bar codes from yeast pools collected immediately before and after elutriation were amplified and differentially labeled with Cy3 (before) and Cy5 (after) and hybridized to bar code microarrays. Green indicates depletion by elutriation (lge cells) while red indicates enrichment (whi cells). (B) Elutriation bar code microarrays experiments were performed with different rich (glucose) (Cook et al. 2008) or poor (glycerol, ethanol) carbon sources. Fitness experiments were performed in the presence of mating pheromone. A number of pheromone pathway gene deletions display a large cell size in the absence of pheromone, and in a manner dependent on the carbon source. The degree of enrichment (small = whi; pheromone-resistant; red) or depletion (large = lge; pheromone sensitive; green) for both upstream and downstream bar codes is expressed as log2(elutriated/total) or log2(treated/untreated) and is indicated by the color bar. Gray indicates no available data either due to insufficient signal or the absence of the strain from the deletion pool. (C) Schematic of the mating pathway. The cell size of gene deletions, relative to the deletion pool, is indicated by the text color: lge (green), whi (red), wild type (black), and no viable data (gray). gpa1Δ and dig1Δ (red asterisks) were both found to be lge; however, they displayed the incorrect pheromone sensitivity/resistance phenotype, likely due to secondary mutations, and were thus excluded.