Figure 6.
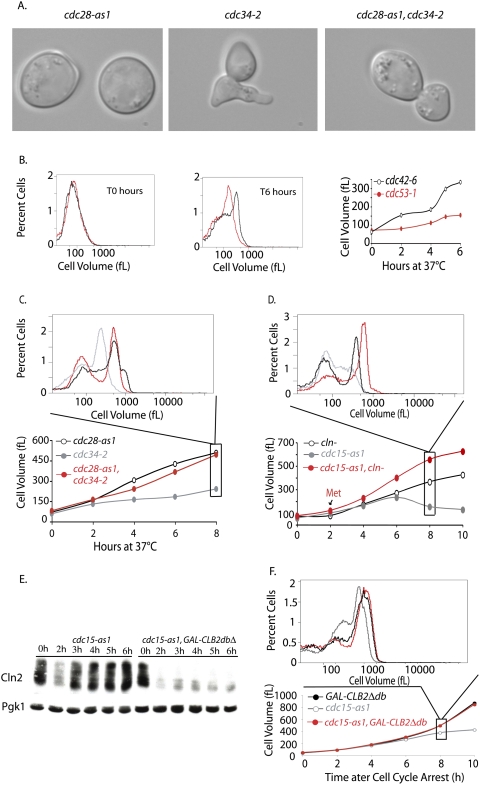
The limited growth of SCF and MEN mutants is due to Cln-CDK activity. (A,C) cdc34-2 (A1467), cdc28-as1 (A4370), and cdc34-2 cdc28-as1 (A17188) cells were shifted to 37°C (0 h time point). Single mutants were treated with 5 μM 1-NM-PP1 inhibitor at the time of temperature shift. The double mutant was grown for 2 h at 37°C, by which time ∼80% of the double mutants had budded, which was followed by addition of CDK inhibitor. (A) Micrographs of mutants after 4 h of growth at 37°C. (B) Comparison of the growth of cdc42-6 (A18221) and cdc53-1 (A1469) mutants. Exponentially growing cells were shifted from room temperature to 37°C, and growth was monitored using a Coulter counter. Cell volume distribution for both strains is shown for the T0 h and T6 h time points, and the mode cell volume for the complete time course is plotted. (C) Inactivation of Cdc28 rescues the growth inhibition of cdc34-2 mutants. Cell size distributions were measured at the indicated time points after shift to 37°C. The histogram above displays the cell volume distribution 8 h after temperature shift. Cells were grown as described in A. (D) Depletion of Cln-CDKs allows cdc15-2 mutants to grow in size. MET-CLN2 cln3Δ cln1Δ (A2624; red circles), cdc15-as1 (A17872; gray circles), and MET-CLN2 cln3Δ cln1Δ cdc15-as1 (A18316; black circles) cells were grown to exponential phase in synthetic complete medium lacking methionine. Cells were transferred into YEPD at 37°C in the presence of 10 μM 1-N-PP1 inhibitor to inhibit the cdc15-as1 allele (0 h time point). The single mutants were treated with 8 mM methionine at the time of temperature shift and cdc15-as1 inhibition, to repress Cln2 production. The double mutant was grown in the absence of methionine for 2 h to allow cells to accumulate in the cdc15-as1 block prior to methionine addition. Subsequently, 8 mM methionine was readded every 2 h to all cultures. Cell size distributions were measured at the indicated time points after shift to 37°C. The histogram above displays the cell volume distribution 8 h after temperature shift. (E) CLNs accumulate in cdc15-as1-arrested cells, but not in cdc15-as1-arrested cells overexpressing CLB2dbΔ. cdc15-as1 (A18982) and cdc15-as1 GAL-CLB2dbΔ (A18986) strains, both expressing a tagged version of CLN2 (Cln2-3xHA), were pregrown at 30°C in YPE supplemented with 2% raffinose. At time 0 h, 10 μM 1-N-PP1 inhibitor and 1% galactose were added to both strains to inhibit cdc15-as1 and to induce CLB2dbΔ, respectively. Samples were withdrawn at the indicated times. Samples were run by 10% SDS-PAGE and prior to transfer to a membrane and blotting. The membrane was blotted with anti-HA antibody to detect Cln2-3X-HA. The same membrane was stripped and reprobed with anti-Pgk1 antibody. (F) Expression of a stable form of Clb2 rescues the growth phenotype of MEN mutants. cdc15-as1 (A17873), GAL-CLB2 dbΔ (A17872), and cdc15-as1 GAL-CLB2 dbΔ double mutant (A17874) were arrested in YPE supplemented with 2% rafinose media with α-factor for 90 min after which 1% galactose was added to induce the expression of Clb2Δdb. Cells were further incubated with α-factor for 45 min and were released into fresh YPE media supplemented with 2% raffinose and 1% galactose at 30°C. At this time, Cdc15-as1 inhibitor, 1-N-PP1, was added to all strains at 10 μM concentration. The volume of all strains was monitored to 10 h. The mode cell volume for each time point is plotted: cdc15-as1 (gray circles), GAL-CLB2dbΔ (black circles), and double mutant (red circles).