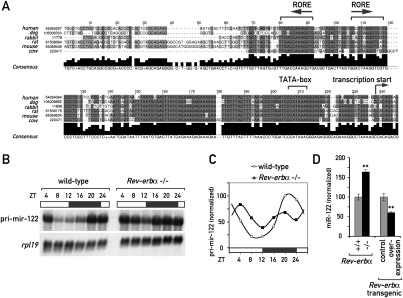
Figure 2.
REV-ERBα is involved in circadian control of the miR-122 locus. (A) Alignment of the genomic sequence upstream of the predicted transcriptional start site of pri-mir-122 in six mammalian species (extracted from the University of California at Santa Cruz alignment; see Supplemental Fig. 3). The predicted ROREs, TATA-box, and transcriptional start site are indicated. (B) Northern blot analysis of pri-mir-122 in total RNA samples from Rev-erbα knockout and littermate control mice sacrificed at the indicated ZT values around the clock. For each time point, an RNA pool of three female mice was used. (C) Quantification of the Northern blot shown in B; values are pri-mir-122 normalized to rpl19. (D) miR-122 levels in total liver RNA from individual animals (mixed ZTs) of the indicated genotypes were quantified by Northern blot (data not shown). Control animals were set to 100%. Data are mean ± SEM (n = 36 for Rev-erbα−/− vs. Rev-erbα+/+ and n = 18 for REV-ERBα overexpression vs. control); (**) P < 5*10−5 (two-tailed Student's t-test).