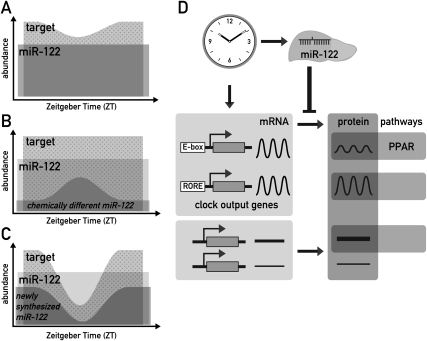
Figure 7.
Models for how miR-122 could impart on circadian gene expression of its targets. (A) Even constant miR-122 levels (dark gray) could shape the circadian rhythm of a target (light gray) by constantly repressing basal levels of translation (represented by the overlap of the two areas). Only the amount of target mRNA represented by the dotted area would be available for translation into protein. As shown in the cartoon, this mechanism could increase the amplitude of cycling and convert a low-amplitude mRNA rhythm into a higher amplitude protein rhythm. In addition, this regulation could confer robustness to low protein expression levels in the trough, as described in the Discussion. The mechanism depicted in this cartoon would require a high affinity of the miRNA–target interaction and an excess of targets over the miRNA. Considering that miR-122 probably has hundreds of targets, of which many contain several seed sites, this assumption is quite plausible even for this highly abundant miRNA (B) Conceivably, chemically distinct, short-lived miR-122 subpopulations (dark gray) could exist within the pool of bulk miR-122. If these distinct miR-122 species also had specific functional properties, this speculative model would imply that target mRNAs would be subject to circadian repression. Consequently, the transcript available to produce protein (dotted area) would show circadian oscillations. (C) Conceivably, newly assembled RISC complexes could immediately get committed to their target mRNAs and remain stably associated with them, as described in the Discussion. The availability of such newly assembled miR-122 RISC would be expected to closely follow circadian miR-122 production (dark gray). If targets are transcribed circadianly as well, the phase relationship of the two rhythmic processes will determine to what extent a target will encounter miR-122 RISCs in the cell, and what influence this has on the circadian amplitude, magnitude, and phase of the produced protein (dotted area). As in A, this model would demand that the miRISC–mRNA affinity be high and that the targets are in excess. (D) Model for the integration of miR-122 in circadian hepatic gene expression. MiR-122 is depicted as a tissue-specific modulator of circadian output genes, with PPAR-dependent regulation of gene expression as one of the regulated output pathways.