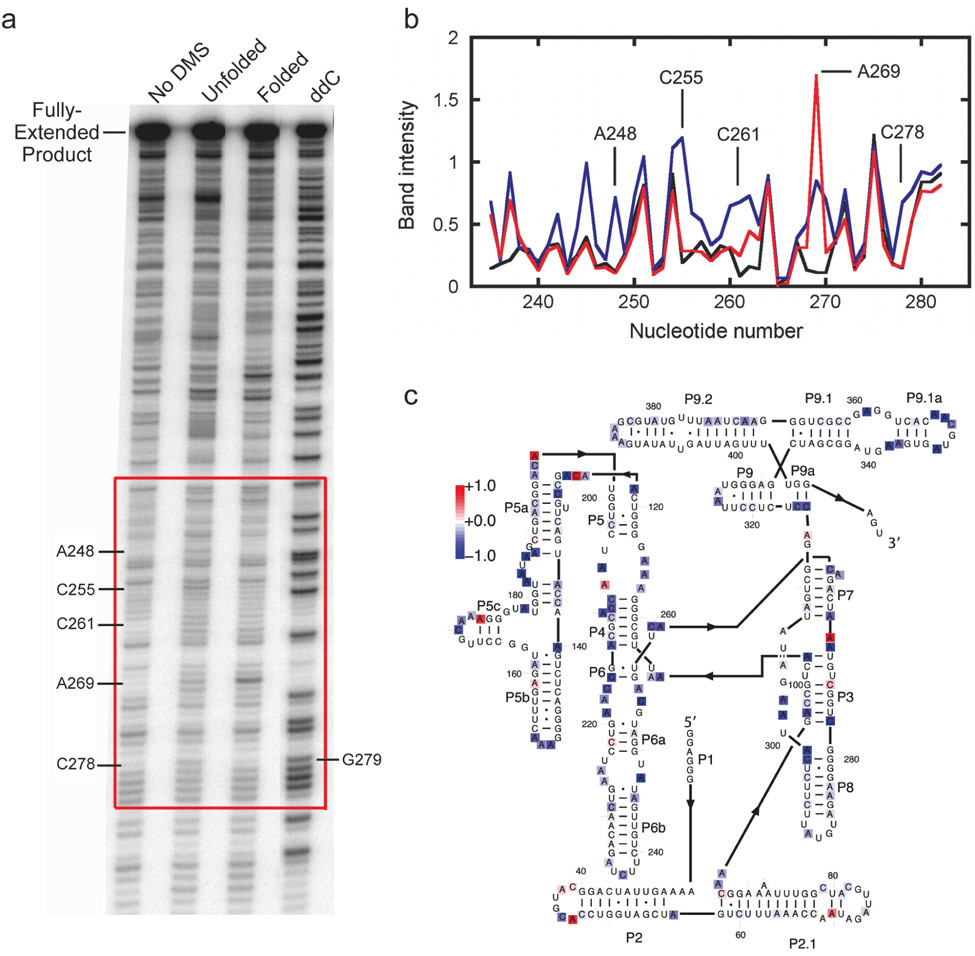
Figure 3. Quantitative analysis of DMS footprinting to monitor RNA folding.
This experiment monitors Mg2+ dependent folding of the Tetrahymena group I ribozyme53. a, DMS footprinting gel. The DMS exposure time for this experiment was relatively short, 1 min at 25 °C with 0.67% DMS, such that the background from RNase degradation and/or pauses by RT is significant relative to the DMS-dependent signal (bands in lane marked ‘No DMS’). However, DMS-dependent bands are readily observed, as indicated by nucleotide labels. The label at the right, G279, is included to illustrate that fragments indicating DMS modification migrate more rapidly by one nucleotide than the corresponding fragments in the sequencing lanes. b, Analysis using SAFA of the region of panel a indicated by the red square. Band intensity was determined by dividing the integrated intensity of each band by the corresponding intensity for the fully-extended primer (determined by boxing this band using Image Quant), and multiplying this by 1000. Thus, an intensity value of 1 means that the indicated band was 0.1% as intense as the fully-extended product, or for the DMS-dependent bands, that roughly 0.1% of the RNA was methylated at this position. c, Comparison of the DMS protection patterns for the folded and unfolded RNAs. Values were obtained by subtracting the intensity values in the unfolded form from those representing the folded form, so that a negative value (colored blue as indicated by the scale bar) represents a nucleotide that is protected in the folded RNA relative to the unfolded RNA, whereas a nucleotide colored red represents a nucleotide that displays enhanced reactivity in the folded RNA.