Abstract
The main function of the nuclear lamina, an intermediate filament meshwork lying primarily beneath the inner nuclear membrane, is to provide structural scaffolding for the cell nucleus. However, the lamina also serves other functions, such as having a role in chromatin organization, connecting the nucleus to the cytoplasm, gene transcription, and mitosis. In somatic cells, the main protein constituents of the nuclear lamina are lamins A, C, B1, and B2. Interest in the nuclear lamins increased dramatically in recent years with the realization that mutations in LMNA, the gene encoding lamins A and C, cause a panoply of human diseases (“laminopathies”), including muscular dystrophy, cardiomyopathy, partial lipodystrophy, and progeroid syndromes. Here, we review the laminopathies and the long strange trip from basic cell biology to therapeutic approaches for these diseases.
The nuclear lamina is an intermediate filament (IF) network composed of proteins called lamins and is part of the nuclear envelope of all somatic cells. For many years, research on the nuclear lamina was the domain of a relatively small group of cell biologists working on nuclear structure and mitosis. In the past decade, however, interest in the nuclear lamina has exploded with the discovery that mutations in the genes encoding lamins and associated nuclear envelope proteins cause a diverse range of human diseases. The mechanisms by which abnormalities in the nuclear lamina cause distinct human diseases (known as laminopathies) involving different tissues and organ systems have remained obscure. This review provides a general introduction to the nuclear lamins, focusing on “lacunae” in our understanding of these proteins. We focus on work in mammals but recognize that important insights regarding lamins have originated from studies of a number of other organisms (in particular, Xenopus, Drosophila, and Caenorhabditis elegans). We also discuss mechanisms by which mutations in lamin A/C (LMNA), the gene that encodes lamins A and C, might cause disease as well as potential therapeutic interventions for laminopathies.
The nuclear envelope and nuclear lamina
The nuclear lamina, an IF network.
The nuclear envelope, which is composed of nuclear membranes, nuclear pore complexes, and the nuclear lamina, separates the nucleus from the cytoplasm. The nuclear lamina is a meshwork of IF proteins known as lamins and is localized primarily on the inner aspect of the inner nuclear membrane (1–4) (Figure 1). In vertebrates, lamins have molecular masses of 60–80 kDa and generally have been divided into two groups, A type and B type, based on differences in isoelectric points (5).
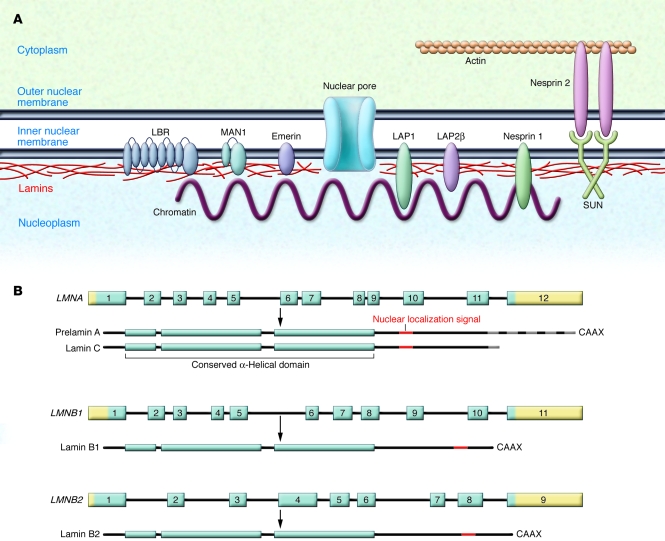
Figure 1. The nuclear lamina.
(A) The nuclear lamina is a meshwork of IFs localized primarily to the nucleoplasmic face of the inner nuclear membrane (shown schematically in red). The lamins interact with several integral proteins of the inner nuclear membrane, including lamin B receptor (LBR), MAN1 (encoded by the LEMD3 gene), emerin, lamina-associated polypeptide 1 (LAP), LAP2β, small nesprin 1 isoforms, and SUNs. SUNs interact with large nesprin 2 isoforms, integral proteins of the outer nuclear membrane, which also interact with actin, linking the nuclear lamina to the cytoskeleton. (B) In humans, 3 genes encode nuclear lamins. LMNA on chromosome 1q21.2 encodes the A-type lamins, with prelamin A and lamin C generated by alternative RNA splicing being the major somatic cell isoforms. Prelamin A has 98 unique amino acids and lamin C 6 unique amino acids at their carboxyl terminus (gray striping). LMNB1 on chromosome 5q23.3–q31.1 encodes lamin B1, and LMNB2 on chromosome 19p13.3 encodes lamin B2, the somatic cell B-type lamins. All the lamins have conserved α-helical rod domains and variable head and tail domains preceding and following the central rod domain. The nuclear localization signals are located in the tail domain (indicated in red). Prelamin A, lamin B1, and lamin B2 have carboxyl-terminal CaaX motifs, a signal for protein farnesylation.
We now know that, in humans, three genes encode nuclear lamins (Figure 1). LMNA on chromosome 1 encodes the A-type lamins, with lamins A and C being the main isoforms in somatic cells (6). Lamins A and C are produced by alternative splicing, and the first 566 amino acids of the two proteins are identical. Lamin C has 6 unique amino acids at its carboxyl terminus, while prelamin A, the precursor of mature lamin A, has 98 unique amino acids. The B-type lamins lamin B1 and lamin B2 are encoded by lamin B1 (LMNB1) on chromosome 5 and lamin B2 (LMNB2) on chromosome 19, respectively (7, 8). B-type lamins are expressed in all somatic cells, whereas lamins A and C are absent from some undifferentiated cells (9, 10). Germ cell–specific transcripts of LMNB1 and LMNA occur as a result of alternative splicing.
Like other IF proteins, lamins have conserved α-helical central rod domains and variable head and tail domains (Figure 1). The basic filament building block is a lamin–lamin dimer. Higher-order polymers are generated from these units, but the precise mechanisms underlying polymer formation are not well understood. It seems that most mammalian lamins can interact with themselves or any other lamin. However, some data show that the strength of binding between different lamins may vary and that A-type and B-type lamins may preferentially polymerize in distinct homopolymers (11, 12). Lamins differ from cytoplasmic IF proteins in that they contain an additional 42 amino acids in their rod domains and have nuclear localization signals in their tail domains.
Lamins not only interact with each other but also with proteins of the inner nuclear membrane, transcription factors, DNA, and chromatin (13) (Figure 1). The organized structure of the nuclear lamina, chromatin, and nuclear envelope in interphase is disrupted during mitosis, when these structures disassemble, allowing chromosome segregation to occur. Nuclear lamina depolymerization during mitosis occurs as a result of phosphorylation of specific amino acids of the lamins (5, 14). Most likely, this is also responsible for the separation of lamins from inner nuclear–membrane proteins and chromatin. During mitosis, the inner nuclear membrane loses its definition and its proteins are resorbed into the ER (15, 16). At the end of mitosis, the nuclear envelope and lamina reassemble in a stepwise manner, with targeting of integral inner nuclear–membrane proteins to chromatin likely to be the first step, followed by pore-complex assembly and lamin polymerization (15–17).
The nuclear lamina was initially thought to mainly provide structural scaffolding for the nuclear envelope, but over the years, numerous studies have implicated lamins in a wide range of functions. In human cells, lamins exist in the nucleoplasm as well as in association with the nuclear envelope (18) and they have been implicated in regulating DNA replication and transcription (19). The nuclear lamins, via interactions with SUNs, inner nuclear–membrane proteins that bind to outer nuclear–membrane proteins known as nesprins, also function as part of a structural network connecting the nucleus to the cytoplasm (20) (Figure 1). How these different functions of lamins relate to disease pathophysiology is not clear.
Posttranslational processing of nuclear lamins.
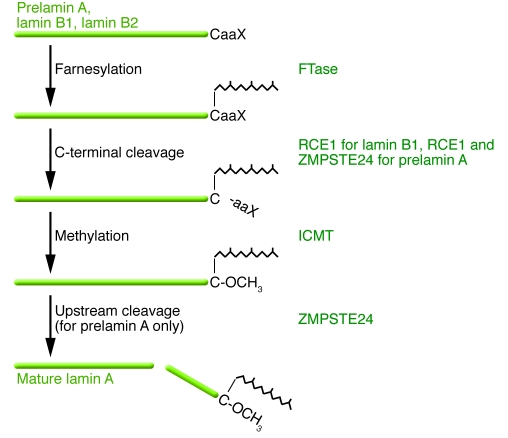
Except for lamin C, mammalian lamins terminate with a CaaX motif (where C is a cysteine, a is often an aliphatic amino acid, and X is one of many different residues). The CaaX motif triggers three sequential enzymatic modifications (21, 22) (Figure 2). First, a 15-carbon farnesyl lipid is added to the cysteine residue by protein farnesyltransferase (FTase) (21, 22). After protein farnesylation, which is one form of protein prenylation, the last three amino acids (the –aaX) are clipped off by an endoprotease specific for prenylated proteins. For lamin B1 and many other CaaX proteins, this proteolytic processing step is carried out by RCE1 homolog, prenyl protein peptidase (RCE1) (23). For prelamin A, this step is likely a redundant function of RCE1 and zinc metallopeptidase, STE24 homolog (ZMPSTE24) (24–26). Next, the newly exposed farnesylcysteine is methylated by a membrane methyltransferase, isoprenylcysteine carboxyl methyltransferase (ICMT) (27, 28). B-type lamins undergo no further modifications and retain a farnesylcysteine α-methyl ester at the carboxyl terminus. In the case of prelamin A, the last 15 amino acids of the protein, including the farnesylcysteine α-methyl ester, are clipped off and degraded, leaving mature lamin A (25, 29, 30) (Figure 2). This final endoproteolytic processing step does not occur in the absence of ZMPSTE24 (24, 31), which very probably carries out this reaction (26). Early studies suggested that the final endoproteolytic processing step required methylation of the farnesylcysteine (24, 32), but follow-up studies revealed that lamin A biogenesis is only modestly affected by the absence of methylation (33). Defining the biochemistry and enzymology of CaaX-motif modifications involved contributions from diverse disciplines, and the history of this topic has been covered elsewhere (21, 22, 25, 34).
Figure 2. Schematic diagram outlining the posttranslational processing of nuclear lamins.
Prelamin A and B-type lamins undergo 3 sequential posttranslational processing steps. First, the cysteine of the carboxyl-terminal CaaX motif is farnesylated by protein FTase. Second, the –aaX is clipped off. For prelamin A, this is likely a redundant activity of RCE1 and ZMPSTE24. Third, the newly exposed carboxyl-terminal farnesylcysteine is methylated by isoprenylcysteine carboxyl methyltransferase (ICMT). Prelamin A undergoes another step in which the carboxyl-terminal 15 amino acids, including the farnesylcysteine methyl ester, are clipped off by ZMPSTE24 and degraded, generating mature lamin A. In the setting of ZMPSTE24 deficiency, the final endoproteolytic cleavage does not occur, leading to the accumulation of a farnesylated and methylated prelamin A. ZMPSTE24 deficiency causes a severe progeroid disorder, RD. In HGPS, an alternative splicing event results in a 50–amino acid deletion in prelamin A, removing the site for the final endoproteolytic cleavage step. Thus, mature lamin A cannot be produced, and cells accumulate a mutant prelamin A that terminates with a farnesylcysteine α-methyl ester.
Why does nature go to the trouble of modifying the CaaX motif of prelamin A, given that the carboxyl terminus of this protein is clipped off and degraded? We believe that the most likely explanation is that the CaaX-motif modifications render prelamin A more hydrophobic, facilitating its targeting to the inner nuclear membrane, where ZMPSTE24 likely releases mature lamin A (35). Farnesylation and methylation increase the hydrophobicity of peptides (36), and Hennekes and Nigg (37) showed that farnesylated prelamin A peptides bind to membrane fractions. The idea that the final cleavage step in lamin A biogenesis occurs at the nuclear lamina was supported by Ottaviano and Gerace (38), who found that a substantial fraction of newly synthesized prelamin A is incorporated into a Triton X-100–insoluble fraction, presumably the nuclear lamina, before its conversion to mature lamin A. Further, a very recent study showed that ZMPSTE24 is located both at the inner nuclear membrane and the ER but suggested that the nucleus is the major site of prelamin A processing (39). However, at this point, no one can exclude the possibility that some prelamin A undergoes processing in the ER.
While prelamin A processing could assist in the delivery of lamin A to the nuclear rim, the processing steps may not be absolutely essential for this process. Michael Sinensky’s group found that a “mature lamin A” variant (where lamin A is synthesized directly, bypassing all processing steps) reaches the nuclear periphery normally (40). Hennekes and Nigg (37) also found that mature lamin A reaches the nuclear periphery but with delayed kinetics.
Diseases related to the nuclear lamina
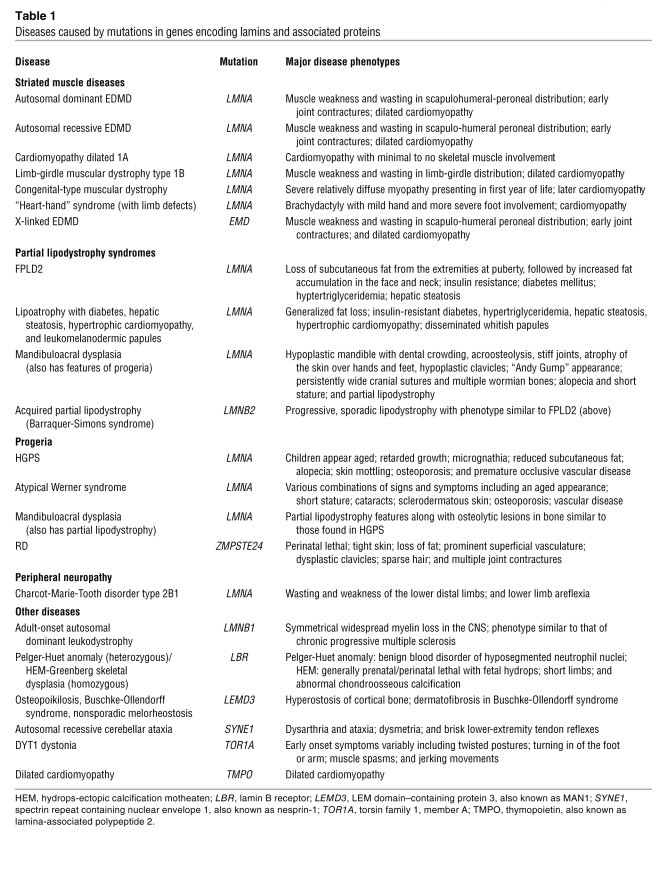
Since 1999, at least 12 disorders have been linked to LMNA mutations. Several of these are probably the same basic disease with variations in severity or extent of organ system involvement. Human diseases also have been associated with mutations in ZMPSTE24, LMNB1, and LMNB2 and mutations in genes encoding proteins that interact with lamins (Table 1).
Table 1 .
Diseases caused by mutations in genes encoding lamins and associated proteins
LMNA.
There are two forms of Emery-Dreifuss muscular dystrophy (EDMD), inherited in either an X-linked or autosomal manner. They are characterized by a triad of muscle weakness and wasting in a scapulohumeral-peroneal distribution; early contractures of the elbows, ankles, and posterior neck; and, most significantly, cardiomyopathy. Symptoms generally appear in the first decade of life, with contractures often the first manifestation. Slowly progressive muscle weakness and wasting usually begin during the second decade of life. Cardiomyopathy occurs in nearly all cases, with the initial presentation usually being a block in atrioventricular conduction followed by left ventricular dilation and congestive heart failure.
In 1994, Bione et al. (41) identified emerin (EMD) as the gene on chromosome Xq28 that is mutated in X-linked EDMD. EMD encodes emerin, an integral protein that was localized to the inner nuclear membrane (42, 43). Later, it was shown that emerin binds directly to A-type lamins (44). In 1999, Bonne et al. (45) demonstrated that mutations in LMNA cause autosomal dominant EDMD. LMNA mutations have also been identified in cases of autosomal recessive EDMD (46). Soon thereafter, other studies showed that LMNA mutations cause dilated cardiomyopathy with conduction defect type 1 (47) and limb-girdle muscular dystrophy type 1B (48). Patients with these autosomal dominant disorders have dilated cardiomyopathy but either little skeletal muscle involvement or involvement in a limb-girdle distribution. These disorders, traditionally classified separately based on clinical criteria, can be caused by the same LMNA mutation and can even occur within the same family (49). Hence, autosomal EDMD, limb-girdle muscular dystrophy type 1B, and dilated cardiomyopathy with conduction defect type 1 can be considered variants of the same disease — with dilated cardiomyopathy and variable skeletal muscle involvement. More recently, LMNA mutations have been shown to cause a congenital muscular dystrophy (50) and “heart-hand” syndrome (51), expanding the range of muscle diseases linked to mutant A-type lamins. Most LMNA mutations causing striated muscle diseases change amino acid residues in lamins A and C, and these amino acid changes can be located throughout the length of these proteins (52). Short deletions, truncating mutations, splicing mutations, and haploinsufficiency can also cause muscle disease (45, 52).
Dunnigan-type familial partial lipodystrophy (FPLD2), also known as Dunnigan-Köbberling syndrome, is an autosomal dominant disorder leading to peripheral loss of adipose tissue. Patients are born with normal fat distribution, but at puberty they develop loss of subcutaneous adipose tissue in the extremities, which produces a muscular appearance, followed by increased adipose tissue accumulation in the face and neck. Visceral fat depots are unaffected or increased. Metabolic manifestations of FPLD2 are insulin resistance, frequently leading to type II diabetes mellitus, and hypertriglyceridemia. These metabolic abnormalities are associated with premature atherosclerosis in some subjects (53). Nearly all individuals with FPLD2 develop hepatic steatosis and may be at increased risk for steatohepatitis (54). Although the disease is autosomal dominant and affects both sexes, the phenotype is easier to discern and may be more severe in women, so there are more female probands (55, 56).
Cao and Hegele (57) and Shackleton et al. (58) reported in 2000 that FPLD2 is caused by mutations in LMNA. Speckman et al. (59) and Vigouroux et al. (55) soon reported additional FPLD2 patients with LMNA mutations. Rare patients with LMNA mutations have been reported that have lipodystrophy syndromes somewhat different than typical FPLD2 in combination with cardiac, skeletal muscle, and other abnormalities (60–62).
While LMNA mutations causing muscle diseases are scattered throughout LMNA, approximately 90% of mutations causing FPLD2 are located in exon 8, most commonly within codons 482 and 486 (55, 57–59). Exon 8 encodes a globular domain common to lamins A and C that forms an immunoglobulin-like fold (63, 64). Myopathy-causing missense mutations in this same domain disrupt the three-dimensional structure of the fold. In contrast, FPLD2 mutations do not disrupt the structure of the domain but alter the charge of solvent-exposed surfaces (63, 64). Mutations in exon 11 of LMNA (in particular codons 582 and 584), a region unique to lamin A, also cause FPLD2, but less frequently (55, 59).
Hutchinson-Gilford progeria syndrome (HGPS) is a rare pediatric progeroid syndrome that has captivated the news media, clinicians, and basic scientists. Children with HGPS appear aged and exhibit retarded growth, micrognathia, reduced subcutaneous fat, alopecia, skin mottling, osteogenic lesions in bone, and osteoporosis. Patients have an increased susceptibility to arterial occlusions, the pathogenesis of which is unclear. The generally held assumption has been that the vascular occlusions are due to atherosclerosis. However, plasma cholesterol levels are normal in individuals with HGPS, and some characteristics of the vessel wall biology in individuals with HGPS are different from those of run-of-the-mill atherosclerosis (65, 66). Thus, it is possible, and perhaps likely, that the vascular occlusions in HGPS are not due to classical atherosclerosis.
HGPS is caused by de novo point mutations that interfere with conversion of farnesyl–prelamin A to mature lamin A (67, 68). The most common mutation is a point mutation in exon 11 (67, 68) that does not alter an amino acid (G608G) but optimizes an alternative splice donor site, resulting in an in-frame deletion of 50 amino acids near the carboxyl terminus of prelamin A (67, 68). Other LMNA point mutations associated with HGPS promote the same aberrant splicing event (67, 69). The 50–amino acid deletion leaves the CaaX motif intact; thus, the mutant prelamin A, generally called progerin, undergoes farnesylation and carboxyl methylation (70). However, the site for the final ZMPSTE24-mediated endoproteolytic cleavage is eliminated (25), preventing further processing to lamin A (71) (Figure 2). Most farnesylated progerin is targeted to the nuclear envelope, although a portion seems to be located in the nucleoplasm (72).
Although typical HGPS almost always involves the accumulation of farnesylated progerin, very similar progeroid syndromes can be caused by missense mutations in LMNA that are not known to be associated with defective prelamin A processing. For example, Verstraeten et al. (73) showed that compound heterozygosity for T28M and M540T mutations caused a progeroid syndrome similar to HGPS. In addition, mandibuloacral dysplasia, a progeroid disorder characterized by lipodystrophy and various bony abnormalities, can be caused by a homozygous R527H mutation (74). How these missense mutations lead to progeroid syndromes requires more investigation.
LMNA mutations can also affect peripheral nerves. A homozygous R298C substitution in lamin A and lamin C has been identified in Algerian families with Charcot-Marie-Tooth disease type 2B1 (75). Affected subjects have weakness and wasting of the distal lower limb muscles, and there is areflexia in the lower limbs. Motor nerve conduction velocities are normal or only slightly reduced in this axonal (nondemyelinating) neuropathy. Subjects with LMNA haploinsufficiency sometimes have axonal neuropathy along with muscular dystrophy/cardiomyopathy (76, 77). Virtually nothing is known about how alterations in A-type lamins cause this type of neuropathy.
The only reported human with complete deficiency of A-type lamins (due to homozygous nonsense mutations in LMNA) exhibited perinatal lethality (78). The preterm infant was smaller than expected for gestational age, died of respiratory failure immediately after birth, and had a slightly dysmorphic face with retrognathia, severe contractures of the limbs, contractures of the fingers and the toes, fractures of the femur and arm, and generalized muscular dystrophy. Hence, some A-type lamin function or functions appear to be necessary for postnatal survival in humans.
ZMPSTE24.
Restrictive dermopathy (RD), a perinatal-lethal progeroid syndrome, is caused by loss of ZMPSTE24 and is associated with the accumulation of farnesylated prelamin A (Figure 2). RD is characterized by tight skin, loss of fat, prominent superficial vasculature, dysplastic clavicles, sparse hair, and multiple joint contractures. Navarro et al. (79) initially proposed that RD was caused by heterozygous mutation of ZMPSTE24 in association with a mutation in another gene. Ultimately, Moulson et al. (80) and Navarro et al. (81) showed that RD results from homozygous loss of ZMPSTE24. Progeroid syndromes less severe than RD, in some cases classified as mandibuloacral dysplasia, can be caused by ZMPSTE24 missense mutations associated with residual enzymatic function (82, 83). Also, one patient with homozygous loss of ZMPSTE24 exhibited phenotypes milder than those in typical RD patients (84); that subject was heterozygous for a truncating mutation in LMNA that eliminated the CaaX motif of prelamin A. This genetic observation strongly suggests that the farnesylated form of prelamin A is responsible for the disease phenotypes of RD.
LMNB1 and LMNB2.
Only two disorders have thus far been linked to modifications in the genes that encode the B-type lamins. First, Padiath et al. (85) identified an LMNB1 duplication in four families with adult-onset autosomal dominant leukodystrophy, a slowly progressive and fatal disorder characterized by pyramidal and cerebellar dysfunction and symmetrical demyelination of the CNS. Overexpression of lamin B1 in the eye of Drosophila causes a degenerative phenotype (85). Second, Hegele’s group (86) identified three heterozygous LMNB2 point mutations, two causing amino acid substitutions, in nine patients with acquired partial lipodystrophy (also known as Barraquer-Simons syndrome). Because these mutations were rare in a control population, the authors suggested that they might be etiologically important in the pathogenesis of acquired partial lipodystrophy. However, no functional studies were performed. Thus far, there have been no associations between human diseases and loss-of-function mutations in either LMNB1 or LMNB2, suggesting either that these proteins serve redundant functions or that loss of these genes causes embryonic lethality. The latter explanation seems more likely, given that Lmnb1 deficiency in mice is perinatal lethal (87).
Abnormal nuclear morphology in laminopathies
To gain insights into the pathogenesis of laminopathies, many investigators began by looking at the most obvious cellular abnormality — misshapen cell nuclei. Indeed, during the past few years, alterations in nuclear morphology induced by lamin abnormalities have received considerable attention. Most studies on how mutant lamins affect nuclear morphology have involved dermal fibroblasts from human patients, fibroblasts from mice with targeted Lmna mutations, or cultured cells that overexpress a mutant lamin protein. Since the initial reports (using fibroblasts from Lmna–/– mice, transfected cells expressing A-type lamin variants detected in individuals with EDMD/cardiomyopathy and FPLD2, and fibroblasts from human subjects with FPLD2; refs. 88–91), there have been many others that have covered virtually all the laminopathies and been reviewed elsewhere (25, 52, 92). The consensus from such studies is that when A-type lamins are absent from cells that normally express them, nuclei are irregular in shape with herniations of the nuclear envelope, nuclear pore complexes cluster slightly, B-type lamins are lost from one nuclear pole, and emerin mislocalizes from the inner nuclear membrane to the bulk ER. Numerous studies (reviewed in refs. 25 and 52) have reported morphological alterations in cells expressing A-type lamin variants, which, depending on the cell type or mutation, have been described as lobulation (also known as blebbing) of the nuclear envelope, honeycombing of the lamina, increased nuclear surface area, thickening of the nuclear lamina, aberrant intranuclear foci of lamins with a decrease at the nuclear periphery, loss of peripheral heterochromatin, and aberrant clustering of nuclear pore complexes. Partial mislocalization of emerin is also observed in cells expressing some A-type lamin variants. Some variants with amino acid substitutions have altered dynamics in the nucleus compared with wild-type lamin A. Cells lacking A-type lamins also exhibit defective nuclear mechanics, and some A-type lamin variants cause increased susceptibility to heat shock. Nuclei of Zmpste24-deficient mouse fibroblasts are misshapen, with blebs and herniations of heterochromatin. Fibroblasts from lamin B1–deficient mice have severely abnormal nuclear morphology with multiple nuclear blebs. Overexpression of B-type lamins induces nuclear membrane growth and the formation of abnormal intranuclear membrane structures, which appear to depend on the presence of a CaaX motif.
Despite numerous reports of abnormal nuclear structure caused by defects in A-type lamins, the significance of these morphological abnormalities to the pathogenesis of disease is unclear. Nuclear abnormalities are generally present only in a subset of fibroblasts, and the percentage of misshapen nuclei depends upon culture conditions, cell density, and passage number. There is no clear association of any specific morphological abnormality with any particular disease. Also, structural alterations in the lamins could lead to abnormalities in other nuclear structures, perturbing nuclear functions not directly linked to structural abnormalities in the nuclear envelope. In any case, the presence of morphological alterations suggests that the mechanical properties of the nuclear lamina are altered, and indeed this has been demonstrated clearly in cells lacking A-type lamins (93, 94). Nuclear morphology in cells expressing either farnesylated prelamin A or progerin can be improved by drugs that block protein farnesylation, and those same drugs improve the whole-animal phenotypes in mice lacking ZMPSTE24 and in mice with HGPS (95–99) (see below). However, in these cases, a cause and effect relationship between improved nuclear shape and improved disease phenotypes has never been established.
Pathogenic mechanisms of laminopathies and potential therapies
How mutations in LMNA cause diverse diseases is poorly understood and is one of the most intriguing riddles in medical genetics. However, recent studies have suggested some insights into the cellular pathogenesis of two LMNA-related disorders, cardiomyopathy and progeria, and those insights have led to studies on potential therapies for these diseases. These studies have relied heavily on the use of genetically modified mouse models (see Sidebar 1 for descriptions of several mouse models relevant to this Review).
Cardiomyopathy.
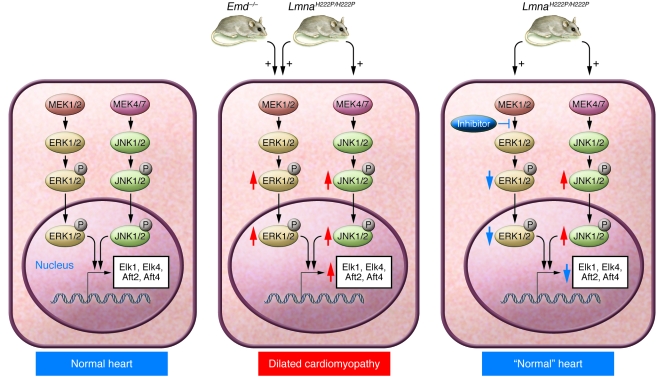
Although cardiomyopathy and muscular dystrophy were the first phenotypes found in human subjects with LMNA mutations, the pathogenic mechanisms underlying these phenotypes have been obscure. To approach this issue, Muchir et al. (100) used microarrays to identify genes and pathways that were perturbed in hearts of LmnaH222P/H222P mice (a mouse model of autosomal EDMD in which a single missense LMNA mutation [H222P] known to cause autosomal EDMD in humans is introduced into Lmna). They detected activation of the ERK and JNK branches of the MAPK signaling cascade, and these changes occurred prior to the onset of histopathologic abnormalities in the heart. Furthermore, expression of mutant forms of lamin A in cultured cells led to activation of ERK and JNK signaling. These findings were consistent with the known alterations in MAPK signaling in cardiomyopathy (101). Muchir et al. (102) further analyzed gene-expression profiles in hearts of Emd-knockout mice, a model of X-linked EDMD, and found a similar molecular signature — activation of ERK and its downstream targets. Thus, MAPK signaling seems relevant to the pathogenesis of heart disease in both X-linked and autosomal dominant EDMD. However, it remains unclear how abnormalities in the nuclear envelope lead to activation of ERK and/or JNK.

Pharmacological inhibitors of MEK (the MAPK kinase that activates ERK) can be administered systemically, and some have been tested as anticancer agents in early-stage human clinical trials. The availability of these agents led Muchir et al. (103) to hypothesize that systemic administration of a MEK inhibitor would prevent dilated cardiomyopathy in LmnaH222P/H222P mice. Beginning at 8 weeks of age, prior to the onset of detectable cardiomyopathy, LmnaH222P/H222P mice were treated with a MEK inhibitor. At 16 weeks of age, nontreated and placebo-treated LmnaH222P/H222P mice manifested increased left ventricular end-systolic and end-diastolic diameters as well as a 30% reduction in ejection fraction. In contrast, hearts of LmnaH222P/H222P mice treated with the MEK inhibitor were indistinguishable from those of wild-type mice. Treatment with a MEK inhibitor also prevented upregulation of genes encoding atrial natriuretic peptides. These mouse studies suggest that ERK inhibition might hold promise for treating cardiomyopathy in human subjects with LMNA and EDM mutations (Figure 3).
Figure 3. Studies from LmnaH222P/H222P knockin mice and Emd-knockout mice suggest that activation of ERK and/or JNK underlies the development of cardiomyopathy.
Cardiomyocytes in normal hearts of wild-type mice exhibit detectable ERK and JNK activation, as judged by low levels of expression of downstream transcription factors such as Elk1, Elk4, Aft2, and Aft4 (left panel). Both ERK and JNK signaling are increased in hearts from mice harboring the H222P point mutation in Lmna, whereas ERK is activated in hearts of Emd-knockout mice (red arrows; middle panel). Phosphorylation and nuclear translocation of ERK and JNK modulate gene expression, leading to dilated cardiomyopathy (middle panel). Currently, it is unclear how alterations in A-type lamins or the loss of emerin lead to the activation of ERK and/or JNK. Studies in LmnaH222P/H222P mice have shown that pharmacological inhibition of MEK, the kinase that phosphorylates ERK, can prevent the development of cardiomyopathy at 16 weeks of age (right panel).
Progeria.
Although no single mechanism has emerged, several studies point to defects in genome maintenance (e.g., defective DNA repair, accumulation of DNA damage, and altered gene expression) as underlying causes of the progeroid syndromes (104). This also seems to be important in progerias associated with ZMPSTE24 deficiency and specific LMNA mutations. Liu et al. (105) reported that ZMPSTE24-deficient mouse fibroblasts were more sensitive to DNA-damaging agents and were slow to repair DNA and proposed that defective DNA repair was linked to genome instability and cell senescence. DNA damage responses are also abnormal in cells from subjects with HGPS, but treatment with a protein FTase inhibitor (FTI), which blocks prenylation of progerin, did not reduce markers of DNA damage (106). Varela et al. (107) used microarray analysis to identify a possible link between ZMPSTE24 deficiency in mice and p53 activation. Also, there was a suggestion that disease phenotypes were less severe in Zmpste24–/–p53–/– mice (107), which would be consistent with the notion that p53 overexpression promotes aging (108). Shumaker et al. (109) showed that histone methylation patterns are altered in HGPS, providing evidence that the lamina interacts with chromatin to modulate heterochromatin and presumably gene transcription. These changes preceded the appearance of abnormally shaped nuclei.
Others have proposed that abnormal nuclear mechanics are an important cellular defect in HGPS. Dahl et al. (110) investigated the mechanical properties of the lamina in HGPS cells and found that the nuclei of HGPS cells exhibited reduced deformability when aspirated with a micropipette. They also found that HGPS nuclei were more resistant to disruption by mechanical pressure than the nuclei from wild-type cells. Verstraeten et al. (111) showed that cultured dermal fibroblasts from patients with HGPS developed progressively stiffer nuclei with increasing passage number and exhibited decreased viability under repetitive mechanical strain as well as attenuated wound healing. FTIs reversed the nuclear stiffness phenotype and accelerated the wound-healing response in fibroblasts from subjects with HGPS and healthy controls but did not restore sensitivity to mechanical strain. While these findings are clearly intriguing, there were no obvious mechanisms connecting the defects in nuclear mechanics to other cellular phenotypes or disease phenotypes.
Signaling pathways required for maintaining normal stem cell function appear to be perturbed in cells expressing progerin or high levels of unprocessed prelamin A. Scaffidi and Misteli (112) showed that the expression of progerin activates downstream effectors of the Notch signaling pathway and alters the differentiation potential of mesenchymal stem cells. Espada et al. (113) showed that ZMPSTE24 deficiency caused an alteration in the number and proliferative capacity of epidermal stem cells, with alterations in molecular signaling pathways implicated in the regulation of stem cells, such as Wnt and microphthalmia transcription factor. These studies demonstrate a potential link between stem cell dysfunction and progeria, but the precise contribution of stem cells to the pathophysiology of progeria remains to be established.
In considering the absence of disease phenotypes in Zmpste24–/–Lmna+/– mice, Fong et al. (95) hypothesized that the farnesylated form of prelamin A might be the molecular culprit in progeria. They further reasoned that blocking protein farnesylation might interfere with progerin targeting to the nuclear periphery, potentially reducing its toxic effects. In support of this hypothesis, Yang et al. (72) showed that FTI treatment mislocalized progerin away from the nuclear periphery and reduced the frequency of misshapen nuclei in LmnaHG/+ fibroblasts (i.e., fibroblasts from mice in which a mutant Lmna allele, LmnaHG, encodes progerin exclusively). Shortly thereafter, Toth et al. (114) showed that an FTI reduced the frequency of misshapen nuclei in cultured fibroblasts from humans with HGPS and ZMPSTE24 deficiency as well as in fibroblasts from Zmpste24–/– mice (114). Several other laboratories reported the same basic findings, some using complementary approaches and different systems (115–118). In the studies by Yang et al. (72) and Toth et al. (114), blockade of protein farnesylation was substantial, as farnesylation of human DnaJ homolog-2 (HDJ-2) (an unrelated CaaX protein) was largely blocked and nonfarnesylated prelamin A accumulated in cells. FTI treatment also reduced levels of lamin A and prelamin A (114), suggesting that the blockade of protein farnesylation might reduce the stability of prelamin A.
The next step was to examine whether an FTI might ameliorate disease in mouse models of progeria. Fong and coworkers (96) found that systemic administration of an FTI improved body weight curves in both male and female Zmpste24–/– mice, although the drug also led to weight loss in wild-type mice. FTI administration also improved survival, improved grip strength performance, and reduced the number of rib fractures. However, FTI-treated Zmpste24–/– mice still had profound disease phenotypes and succumbed to the progeroid disease. In these studies, only 10%–50% of the HDJ-2 in tail extracts was nonfarnesylated (similar to levels observed in the testing of FTIs as anticancer agents). The FTI also led to the appearance of nonfarnesylated prelamin A in tissue extracts but to a lower extent than in cell culture experiments (96). LmnaHG/+ mice treated with an FTI also exhibited improvements in body weight curves, weights of fat depots, bone fractures, and bone mineralization (97, 99). However, as in Zmpste24–/– mice, improvements in disease phenotypes fell far short of a cure. In these studies, more than 50% of HDJ-2 in livers of FTI-treated mice was nonfarnesylated, and small amounts of nonfarnesylated prelamin A accumulated in tissues (97, 99). Increasing FTI doses led to greater effects on farnesylation, but survival was adversely affected, presumably because of drug toxicity (99).
The efficacy of FTIs in reducing the number of cells with misshapen nuclei (72, 114) and in ameliorating disease phenotypes in mouse models of progeria (96, 97, 99) prompted an open-label trial of an FTI (lonafarnib) in children with HGPS (119). The trial has been ongoing for more than a year, but neither clinical outcomes nor evidence regarding the in vivo blockade of protein farnesylation are available. In the human trial, lonafarnib is being administered to children with advanced disease phenotypes, a substantial difference from the mouse experiments, where the drug therapy was initiated prior to development of significant disease.
The improvements in nuclear shape in FTI-treated HGPS fibroblasts have in some cases been quite striking (72, 116). However, the benefits of FTIs in LmnaHG/+ mice, although highly statistically significant, have been less dramatic. One possible explanation is that the degree of inhibition of protein FTase was less than complete in the mice. Another possibility is that prelamin A can be alternately prenylated by geranylgeranyltransferase-I (GGTase-I) when FTase is blocked. The latter possibility is very plausible, as lamins terminate with methionine and other CaaX proteins terminating with methionine can be geranylgeranylated when FTase is blocked (120). Varela et al. (98) used mass spectrometry (MALDI-TOF) to examine prelamin A and progerin structure in FTI-treated cells and uncovered strong evidence that these proteins are geranylgeranylated in FTI-treated fibroblasts. Whether alternate geranylgeranylation of prelamin A also occurred in FTI-treated mice was never investigated. Varela et al. (98) also reported that a combination of a GGTase-I inhibitor and an FTI increased prelamin A accumulation in cultured fibroblasts, again consistent with alternate prenylation. On the other hand, another group found that an FTI retards the electrophoretic migration of prelamin A, raising the possibility that the extent of alternate prenylation may be limited (or that the FTIs that were used are effective in blocking protein geranylgeranylation) (114). More studies are clearly required to define the extent of alternate prenylation in vivo in the setting of FTI therapy and to explore its potential relevance to the pathogenesis of progeria.
Another possible explanation for the incomplete therapeutic response with an FTI in LmnaHG/+ mice is that it leads to accumulation of another abnormal lamin—nonfarnesylated progerin. If the nonfarnesylated progerin were itself toxic to cells, the benefits of FTI therapy would obviously be limited. To explore the possibility that nonfarnesylated progerin is toxic and capable of eliciting disease, Yang et al. (121) created knockin mice expressing nonfarnesylated progerin (LmnanHG/+). LmnanHG/+ mice are genetically identical to LmnaHG/+ mice, except that the cysteine in the CaaX motif of progerin was changed to serine, eliminating all protein prenylation. LmnanHG/+ mice exhibited the same phenotypes as LmnaHG/+ mice, but they were slightly milder. In addition, fewer LmnanHG/+ fibroblasts contained misshapen nuclei (121). A likely explanation for the milder phenotypes in LmnanHG/+ mice was that steady-state cellular levels of progerin were lower in LmnanHG/+ than LmnaHG/+ mice (121). The milder phenotypes in LmnanHG/+ mice (compared with LmnaHG/+ mice) are consistent with the results of FTI treatment studies (97, 99) and support the idea that inhibiting protein farnesylation could be beneficial. On the other hand, finding that nonfarnesylated progerin caused substantial disease suggested that there could be significant limitations in the FTI treatment strategy for HGPS.
Toth et al. (114) suggested that bisphosphonates, frequently prescribed for osteoporosis, might be useful for treating bone disease in progeria. Nitrogen-containing bisphosphonates bind avidly to bone and block farnesyl diphosphate synthase, an enzyme that produces farnesyl diphosphate (122). Inhibiting synthesis of farnesyl diphosphate blocks protein farnesylation, protein geranylgeranylation, and cholesterol synthesis, and these effects are thought to promote apoptosis in osteoclasts, leading to improved bone density. It is not clear that increased activity of osteoclasts underlies bone disease of progeria (25), and it seems somewhat more probable that dysfunctional osteoblasts are more important for the pathogenesis of disease (24, 25). In any case, Toth et al. (114) showed that one of the nitrogen-containing bisphosphonates, alendronate, inhibited prelamin A processing in wild-type and HGPS fibroblasts, although the blockade of lamin A biogenesis was less than with an FTI. The impact of these drugs on prelamin A processing in vivo (in mouse or humans) is not yet clear. A potential advantage of bisphosphonates is that they would interfere with geranylgeranylation by GGTase-I, if indeed this enzyme were active in the posttranslational modification of prelamin A in vivo. Another advantage of bisphosphonates is that these drugs are concentrated in bone, a tissue that is severely affected in progeria. On the other hand, it seems unlikely that these drugs would be helpful for treating disease phenotypes unrelated to bone, such as lipodystrophy and vascular disease.
Varela et al. (98) tested a combination of a potent nitrogen-containing bisphosphonate and a statin in Zmpste24–/– mice and documented improved survival and improvements in bone abnormalities. The rationale for this drug combination was to inhibit prelamin A farnesylation in a synergistic manner and also to block any alternate geranylgeranylation that might occur in the tissues of mice. Although it is clear that the mice treated with the drug combination exhibited an improvement in disease phenotypes, they did not report whether the drug combination actually affected the prenylation of prelamin A (or any other protein) in the tissues of mice; thus, the mechanism for the observed improvements in disease phenotypes is not yet entirely clear. Varela et al. (98) did not find a beneficial effect of a statin alone or a bisphosphonate alone on the survival of Zmpste24–/– mice, although the number of mice in the latter experiments was small.
Blocking protein prenylation with an FTI represents a “blunt instrument” for the treatment of HGPS, in that FTIs block the posttranslational processing of many CaaX proteins, including the B-type lamins. Similarly, the bisphosphonate/statin combination would inhibit the processing of both farnesylated and geranylgeranylated proteins, at least in bone. A more specific approach would be to identify therapies that interfere with the proximal cause of HGPS — the utilization of the alternate splice donor site in exon 11 of LMNA. Scaffidi and Misteli (123) transfected HGPS fibroblasts with a morpholino oligonucleotide directed against the abnormal splice donor site and found reduced levels of progerin in cells, reduced frequency of misshapen nuclei in fibroblasts, and normalized expression of aberrantly expressed genes. Huang et al. (124) tested an RNA interference approach to reduce levels of progerin in HGPS cells. They identified a short hairpin RNA directed against sequences unique to progerin that reduced progerin transcripts and protein levels by approximately 25%. Despite this modest effect, the frequency of misshapen nuclei was reduced and cell proliferation rates increased. Another strategy was suggested by the absence of disease in mice carrying two Lmna alleles that exclusively produce lamin C (LmnaLCO/LCO mice, so called lamin C–only mice) (125). If lamin A and prelamin A are dispensable, it might make sense to treat HGPS by eliminating all prelamin A transcripts (both for progerin and wild-type prelamin A) with antisense oligonucleotides. Fong et al. (125) identified an antisense oligonucleotide that potently reduced prelamin A transcripts and, when tested in fibroblasts from mice lacking ZMPSTE24, reduced both prelamin A levels and the frequency of misshapen nuclei. Thus far, no laboratory has tested oligonucleotide therapeutics in animal models.
Conclusion: laminopathies as rare disease models of common conditions
Research on nuclear lamins and laminopathies provides a fascinating example of an unexpected intersection between basic cell biology and clinical medicine. During the next few years, we anticipate many more studies on disease pathogenesis and treatment approaches. These studies are important because, although monogenic laminopathies are relatively rare, investigation of these disorders is likely to yield important insights into cellular processes involved in more common forms of cardiomyopathy, metabolic disorders, and physiological aging.
Acknowledgments
We apologize to colleagues in this field that we could not cite all of the important papers due to strict limitations on space and numbers of references. The authors were supported by NIH grants HL76839, CA099506, and AR050200 (to S.G. Young); HL086683 (to L.G. Fong); AR048997, NS059352, and AG025240 (to H.J. Worman); Muscular Dystrophy Association grant MDA4287 (to H.J. Worman); Ellison Medical Research Foundation grant AGSS1678 (to S.G. Young); and March of Dimes grant FY20071012 (to L.G. Fong).
Footnotes
Conflict of interest: H.J. Worman and A. Muchir are inventors on an international patent application filed by the Trustees of Columbia University on MAP kinase inhibition to treat cardiomyopathy.
Nonstandard abbreviations used: EDMD, Emery-Dreifuss muscular dystrophy; FPLD2, Dunnigan-type familial partial lipodystrophy; FTase, farnesyltransferase; FTI, FTase inhibitor; GGTase-I, geranylgeranyltransferase-I; HDJ-2, human DnaJ homolog-2; HGPS, Hutchinson-Gilford progeria syndrome; IF, intermediate filament; RD, restrictive dermopathy; ZMPSTE24, zinc metallopeptidase, STE24 homolog.
Citation for this article: J. Clin. Invest. 119:1825–1836 (2009). doi:10.1172/JCI37679
Antoine Muchir’s present address is: Santhera Pharmaceuticals Ltd., Liestal, Switzerland.
References
- 1.McKeon F.D., Kirschner M.W., Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- 2.Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 3.Goldman A.E., Maul G., Steinert P.M., Yang H.Y., Goldman R.D. Keratin-like proteins that coisolate with intermediate filaments of BHK-21 cells are nuclear lamins. Proc. Natl. Acad. Sci. U. S. A. 1986;83:3839–3843. doi: 10.1073/pnas.83.11.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher D.Z., Chaudhary N., Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. U. S. A. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerace L., Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- 6.Lin F., Worman H.J. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- 7.Biamonti G., et al. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol. Cell. Biol. 1992;12:3499–3506. doi: 10.1128/mcb.12.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin F., Worman H.J. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 1995;27:230–236. doi: 10.1006/geno.1995.1036. [DOI] [PubMed] [Google Scholar]
- 9.Stewart C., Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- 10.Guilly M.N., Bensussan A., Bourge J.F., Bornens M., Courvalin J.C. A human T lymphoblastic cell line lacks lamins A and C. EMBO J. 1987;6:3795–3799. doi: 10.1002/j.1460-2075.1987.tb02715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schirmer E.C., Gerace L. The stability of the nuclear lamina polymer changes with the composition of lamin subtypes according to their individual binding strengths. J. Biol. Chem. 2004;279:42811–42817. doi: 10.1074/jbc.M407705200. [DOI] [PubMed] [Google Scholar]
- 12.Delbarre E., et al. The truncated prelamin A in Hutchinson-Gilford progeria syndrome alters segregation of A-type and B-type lamin homopolymers. Hum. Mol. Genet. 2006;15:1113–1122. doi: 10.1093/hmg/ddl026. [DOI] [PubMed] [Google Scholar]
- 13.Schirmer E.C., Foisner R. Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 2007;313:2167–2179. doi: 10.1016/j.yexcr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Nigg E.A. Assembly and cell cycle dynamics of the nuclear lamina. Semin. Cell Biol. 1992;3:245–253. doi: 10.1016/1043-4682(92)90026-R. [DOI] [PubMed] [Google Scholar]
- 15.Ellenberg J., et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L., Guan T., Gerace L. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol. 1997;137:1199–1210. doi: 10.1083/jcb.137.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhary N., Courvalin J.C. Stepwise reassembly of the nuclear envelope at the end of mitosis. J. Cell Biol. 1993;122:295–306. doi: 10.1083/jcb.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hozak P., Sasseville A.M., Raymond Y., Cook P.R. Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J. Cell. Sci. 1995;108:635–644. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- 19.Dechat T., et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Worman H.J., Gundersen G.G. Here come the SUNs: a nucleocytoskeletal missing link. Trends Cell Biol. 2006;16:67–69. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu. Rev. Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- 22.Zhang F.L., Casey P.J. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 23.Maske C.P., et al. A carboxyl-terminal interaction of lamin B1 is dependent on the CAAX endoprotease Rce1 and carboxymethylation. J. Cell Biol. 2003;162:1223–1232. doi: 10.1083/jcb.200303113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergo M.O., et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. . Proc. Natl. Acad. Sci. U. S. A. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young S.G., Fong L.G., Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—New evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Corrigan D.P., et al. Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem. J. 2005;387:129–138. doi: 10.1042/BJ20041359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke S., Vogel J.P., Deschenes R.J., Stock J. Posttranslational modification of the Ha-ras oncogene protein: Evidence for a third class of protein carboxyl methyltransferases. . Proc. Natl. Acad. Sci. U. S. A. 1988;85:4643–4647. doi: 10.1073/pnas.85.13.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai Q., et al. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J. Biol. Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- 29.Weber K., Plessmann U., Traub P. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 1989;257:411–414. doi: 10.1016/0014-5793(89)81584-4. [DOI] [PubMed] [Google Scholar]
- 30.Beck L.A., Hosick T.J., Sinensky M. Isoprenylation is required for the processing of the lamin A precursor. J. Cell Biol. 1990;110:1489–1499. doi: 10.1083/jcb.110.5.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pendás A.M., et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase–deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 32.Kilic F., et al. In vitro assay and characterization of the farnesylation-dependent prelamin A endoprotease. J. Biol. Chem. 1997;272:5298–5304. doi: 10.1074/jbc.272.8.5298. [DOI] [PubMed] [Google Scholar]
- 33.Young, S.G., Clarke, S., Bergo, M., Philips, M., and Fong, L.G. 2005. Genetic approaches to understanding the physiologic importance of the carboxyl methylation of isoprenylated proteins. In The enzymes. S. Clarke and F. Tamanoi, editors. Academic Press. San Diego, California, USA. 273–301. [DOI] [PubMed] [Google Scholar]
- 34.Young, S.G., Ambroziak, P., Kim, E., and Clarke, S. 2000. Postisoprenylation protein processing: CXXX (CaaX) endoproteases and isoprenylcysteine carboxyl methyltransferase. In The enzymes. F. Tamanoi and D.S. Sigman, editors. Academic Press. San Diego, California, USA. 155–213. [Google Scholar]
- 35.Holtz D., Tanaka R.A., Hartwig J., McKeon F. The CaaX motif of lamin A functions in conjunction with the nuclear localization signal to target assembly to the nuclear envelope. Cell. 1989;59:969–977. doi: 10.1016/0092-8674(89)90753-8. [DOI] [PubMed] [Google Scholar]
- 36.Silvius J.R., l’Heureux F. Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry. 1994;33:3014–3022. doi: 10.1021/bi00176a034. [DOI] [PubMed] [Google Scholar]
- 37.Hennekes H., Nigg E.A. The role of isoprenylation in membrane attachment of nuclear lamins. A single point mutation prevents proteolytic cleavage of the lamin A precursor and confers membrane binding properties. J. Cell. Sci. 1994;107:1019–1029. doi: 10.1242/jcs.107.4.1019. [DOI] [PubMed] [Google Scholar]
- 38.Ottaviano Y., Gerace L. Phosphorylation of the nuclear lamins during interphase and mitosis. J. Biol. Chem. 1985;260:624–632. [PubMed] [Google Scholar]
- 39.Barrowman J., Hamblet C., George C.M., Michaelis S. Analysis of Prelamin a Biogenesis Reveals the Nucleus to be a CaaX Processing Compartment. Mol. Biol. Cell. 2008;19:5398–5408. doi: 10.1091/mbc.E08-07-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lutz R.J., Trujillo M.A., Denham K.S., Wenger L., Sinensky M. Nucleoplasmic localization of prelamin A: Implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc. Natl. Acad. Sci. U. S. A. 1992;89:3000–3004. doi: 10.1073/pnas.89.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bione S., et al. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 1994;8:323–327. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- 42.Nagano A., et al. Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy. Nat. Genet. 1996;12:254–259. doi: 10.1038/ng0396-254. [DOI] [PubMed] [Google Scholar]
- 43.Manilal S., Nguyen T.M., Sewry C.A., Morris G.E. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet. 1996;5:801–808. doi: 10.1093/hmg/5.6.801. [DOI] [PubMed] [Google Scholar]
- 44.Clements L., Manilal S., Love D.R., Morris G.E. Direct interaction between emerin and lamin A. Biochem. Biophys. Res. Commun. 2000;267:709–714. doi: 10.1006/bbrc.1999.2023. [DOI] [PubMed] [Google Scholar]
- 45.Bonne G., et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 46.Raffaele Di Barletta M., et al. Different mutations in the LMNA gene cause autosomal dominant and autosomal recessive Emery-Dreifuss muscular dystrophy. Am. J. Hum. Genet. 2000;66:1407–1412. doi: 10.1086/302869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fatkin D., et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 48.Muchir A., et al. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum. Mol. Genet. 2000;9:1453–1459. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- 49.Brodsky G.L., et al. Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement. Circulation. 2000;101:473–476. doi: 10.1161/01.cir.101.5.473. [DOI] [PubMed] [Google Scholar]
- 50.Quijano-Roy S., et al. De novo LMNA mutations cause a new form of congenital muscular dystrophy. Ann. Neurol. 2008;64:177–186. doi: 10.1002/ana.21417. [DOI] [PubMed] [Google Scholar]
- 51.Renou L., et al. Heart-hand syndrome of Slovenian type: a new kind of laminopathy. J. Med. Genet. 2008;45:666–671. doi: 10.1136/jmg.2008.060020. [DOI] [PubMed] [Google Scholar]
- 52.Worman H.J., Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp. Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hegele R.A. Premature atherosclerosis associated with monogenic insulin resistance. Circulation. 2001;103:2225–2229. doi: 10.1161/01.cir.103.18.2225. [DOI] [PubMed] [Google Scholar]
- 54.Ludtke A., et al. Hepatic steatosis in Dunnigan-type familial partial lipodystrophy. Am. J. Gastroenterol. 2005;100:2218–2224. doi: 10.1111/j.1572-0241.2005.00234.x. [DOI] [PubMed] [Google Scholar]
- 55.Vigouroux C., et al. Lamin A/C gene: sex-determined expression of mutations in Dunnigan-type familial partial lipodystrophy and absence of coding mutations in congenital and acquired generalized lipoatrophy. Diabetes. 2000;49:1958–1962. doi: 10.2337/diabetes.49.11.1958. [DOI] [PubMed] [Google Scholar]
- 56.Garg A. Gender differences in the prevalence of metabolic complications in familial partial lipodystrophy (Dunnigan variety). J. Clin. Endocrinol. Metab. 2000;85:1776–1782. doi: 10.1210/jc.85.5.1776. [DOI] [PubMed] [Google Scholar]
- 57.Cao H., Hegele R.A. Nuclear lamin A/C R482Q mutation in canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- 58.Shackleton S., et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat. Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 59.Speckman R.A., et al. Mutational and haplotype analyses of families with familial partial lipodystrophy (Dunnigan variety) reveal recurrent missense mutations in the globular C-terminal domain of lamin A/C. Am. J. Hum. Genet. 2000;66:1192–1198. doi: 10.1086/302836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caux F., et al. A new clinical condition linked to a novel mutation in lamins A and C with generalized lipoatrophy, insulin-resistant diabetes, disseminated leukomelanodermic papules, liver steatosis, and cardiomyopathy. J. Clin. Endocrinol. Metab. 2003;88:1006–1013. doi: 10.1210/jc.2002-021506. [DOI] [PubMed] [Google Scholar]
- 61.van der Kooi A.J., et al. Lamin A/C mutations with lipodystrophy, cardiac abnormalities, and muscular dystrophy. Neurology. 2002;59:620–623. doi: 10.1212/wnl.59.4.620. [DOI] [PubMed] [Google Scholar]
- 62.Garg A., Speckman R.A., Bowcock A.M. Multisystem dystrophy syndrome due to novel missense mutations in the amino-terminal head and alpha-helical rod domains of the lamin A/C gene. Am. J. Med. 2002;112:549–555. doi: 10.1016/S0002-9343(02)01070-7. [DOI] [PubMed] [Google Scholar]
- 63.Krimm I., et al. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 2002;10:811–823. doi: 10.1016/S0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- 64.Dhe-Paganon S., Werner E.D., Chi Y.-I., Shoelson S.E. Structure of the globular tail of nuclear lamin. J. Biol. Chem. 2002;277:17381–17384. doi: 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- 65.Merideth M.A., et al. Phenotype and course of Hutchinson-Gilford progeria syndrome. N. Engl. J. Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gordon L.B., Harten I.A., Patti M.E., Lichtenstein A.H. Reduced adiponectin and HDL cholesterol without elevated C-reactive protein: clues to the biology of premature atherosclerosis in Hutchinson-Gilford Progeria Syndrome. J. Pediatr. 2005;146:336–341. doi: 10.1016/j.jpeds.2004.10.064. [DOI] [PubMed] [Google Scholar]
- 67.Eriksson M., et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. . Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Sandre-Giovannoli A., et al. Lamin A truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 69.Moulson C.L., et al. Increased progerin expression associated with unusual LMNA mutations causes severe progeroid syndromes. Hum. Mutat. 2007;28:882–889. doi: 10.1002/humu.20536. [DOI] [PubMed] [Google Scholar]
- 70.Dechat T., et al. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4955–4960. doi: 10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goldman R.D., et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang S.H., et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10291–10296. doi: 10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verstraeten V.L., et al. Compound heterozygosity for mutations in LMNA causes a progeria syndrome without prelamin A accumulation. Hum. Mol. Genet. 2006;15:2509–2522. doi: 10.1093/hmg/ddl172. [DOI] [PubMed] [Google Scholar]
- 74.Novelli G., et al. Mandibuloacral dysplasia is caused by a mutation in LMNA-encoding lamin A/C. . Am. J. Hum. Genet. 2002;71:426–431. doi: 10.1086/341908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Sandre-Giovannoli A., et al. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. . Am. J. Hum. Genet. 2002;70:726–736. doi: 10.1086/339274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walter M.C., et al. Deletion of the LMNA initiator codon leading to a neurogenic variant of autosomal dominant Emery-Dreifuss muscular dystrophy. Neuromuscul. Disord. 2005;15:40–44. doi: 10.1016/j.nmd.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 77.Benedetti S., et al. Dominant LMNA mutations can cause combined muscular dystrophy and peripheral neuropathy. J. Neurol. Neurosurg. Psychiatr. 2005;76:1019–1021. doi: 10.1136/jnnp.2004.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Engelen B.G., et al. The lethal phenotype of a homozygous nonsense mutation in the lamin A/C gene. Neurology. 2005;64:374–376. doi: 10.1212/01.WNL.0000149763.15180.00. [DOI] [PubMed] [Google Scholar]
- 79.Navarro C., et al. Lamin A and ZMPSTE24 defects cause nuclear disorganization and identify restrictive dermopathy as a lethal neonatal laminopathy. Hum. Mol. Genet. 2004;13:2493–2503. doi: 10.1093/hmg/ddh265. [DOI] [PubMed] [Google Scholar]
- 80.Moulson C.L., et al. Homozygous and compound heterozygous mutations in ZMPSTE24 cause the laminopathy restrictive dermopathy. J. Invest. Dermatol. 2005;125:913–919. doi: 10.1111/j.0022-202X.2005.23846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Navarro C.L., et al. Loss of ZMPSTE24 (FACE-1) causes autosomal recessive restrictive dermopathy and accumulation of Lamin A precursors. Hum. Mol. Genet. 2005;14:1503–1513. doi: 10.1093/hmg/ddi159. [DOI] [PubMed] [Google Scholar]
- 82.Agarwal A.K., Fryns J.-P., Auchus R.J., Garg A. Zinc metalloproteinase, ZMPSTE24, is mutated in mandibuloacral dysplasia. . Hum. Mol. Genet. 2003;12:1995–2001. doi: 10.1093/hmg/ddg213. [DOI] [PubMed] [Google Scholar]
- 83.Shackleton S., et al. Compound heterozygous ZMPSTE24 mutations reduce prelamin A processing and result in a severe progeroid phenotype. J. Med. Genet. 2005;42:e36. doi: 10.1136/jmg.2004.029751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Denecke J., et al. A homozygous ZMPSTE24 null mutation in combination with a heterozygous mutation in the LMNA gene causes Hutchinson-Gilford progeria syndrome (HGPS): insights into the pathophysiology of HGPS. Hum. Mutat. 2006;27:524–531. doi: 10.1002/humu.20315. [DOI] [PubMed] [Google Scholar]
- 85.Padiath Q.S., et al. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat. Genet. 2006;38:1114–1123. doi: 10.1038/ng1872. [DOI] [PubMed] [Google Scholar]
- 86.Hegele R.A., et al. Sequencing of the reannotated LMNB2 gene reveals novel mutations in patients with acquired partial lipodystrophy. Am. J. Hum. Genet. 2006;79:383–389. doi: 10.1086/505885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vergnes L., Peterfy M., Bergo M.O., Young S.G., Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sullivan T., et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ostlund C., Bonne G., Schwartz K., Worman H.J. Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J. Cell. Sci. 2001;114:4435–4445. doi: 10.1242/jcs.114.24.4435. [DOI] [PubMed] [Google Scholar]
- 90.Raharjo W.H., Enarson P., Sullivan T., Stewart C.L., Burke B. Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery-Dreifuss muscular dystrophy. . J. Cell. Sci. 2001;114:4447–4457. doi: 10.1242/jcs.114.24.4447. [DOI] [PubMed] [Google Scholar]
- 91.Vigouroux C., et al. Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J. Cell. Sci. 2001;114:4459–4468. doi: 10.1242/jcs.114.24.4459. [DOI] [PubMed] [Google Scholar]
- 92.Stewart C.L., Kozlov S., Fong L.G., Young S.G. Mouse models of the laminopathies. Exp. Cell Res. 2007;313:2144–2156. doi: 10.1016/j.yexcr.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lammerding J., et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Broers J.L., et al. Decreased mechanical stiffness in LMNA–/– cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. . Hum. Mol. Genet. 2004;13:2567–2580. doi: 10.1093/hmg/ddh295. [DOI] [PubMed] [Google Scholar]
- 95.Fong L.G., et al. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2004;101:18111–18116. doi: 10.1073/pnas.0408558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fong L., et al. A protein farnesyltransferase inhibitor ameliorates disease in a mouse model of progeria. Science. 2006;311:1621–1623. doi: 10.1126/science.1124875. [DOI] [PubMed] [Google Scholar]
- 97.Yang S.H., et al. Treatment with a protein farnesyltransferase inhibitor improves disease phenotypes in mice with a targeted Hutchinson-Gilford progeria syndrome mutation. J. Clin. Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Varela I., et al. Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat. Med. 2008;14:767–772. doi: 10.1038/nm1786. [DOI] [PubMed] [Google Scholar]
- 99.Yang S.H., Qiao X., Fong L.G., Young S.G. Treatment with a farnesyltransferase inhibitor improves survival in mice with a Hutchinson-Gilford progeria syndrome mutation. Biochim. Biophys. Acta. 2008;1781:36–39. doi: 10.1016/j.bbalip.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Muchir A., et al. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J. Clin. Invest. 2007;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Molkentin J.D. Calcineurin-NFAT signaling regulates the cardiac hypertrophic response in coordination with the MAPKs. Cardiovasc. Res. 2004;63:467–475. doi: 10.1016/j.cardiores.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 102.Muchir A., Pavlidis P., Bonne G., Hayashi Y.K., Worman H.J. Activation of MAPK in hearts of EMD null mice: similarities between mouse models of X-linked and autosomal dominant Emery Dreifuss muscular dystrophy. Hum. Mol. Genet. 2007;16:1884–1895. doi: 10.1093/hmg/ddm137. [DOI] [PubMed] [Google Scholar]
- 103.Muchir A., Shan J., Bonne G., Lehnart S.E., Worman H.J. Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum. Mol. Genet. . 2009;18:241–247. doi: 10.1093/hmg/ddn343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hasty P., Campisi J., Hoeijmakers J., vanSteeg H., Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 105.Liu B., et al. Genomic instability in laminopathy-based premature aging. Nat. Med. 2005;11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 106.Liu Y., Rusinol A., Sinensky M., Wang Y., Zou Y. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J. Cell. Sci. 2006;119:4644–4649. doi: 10.1242/jcs.03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Varela I., et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 108.Tyner S.D., et al. p53 mutant mice that display early ageing-associated phenotytpes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 109.Shumaker D.K., et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dahl K.N., et al. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Verstraeten V.L., Ji J.Y., Cummings K.S., Lee R.T., Lammerding J. Increased mechanosensitivity and nuclear stiffness in Hutchinson-Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell. 2008;7:383–393. doi: 10.1111/j.1474-9726.2008.00382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Scaffidi P., Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat. Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Espada J., et al. Nuclear envelope defects cause stem cell dysfunction in premature-aging mice. J. Cell Biol. 2008;181:27–35. doi: 10.1083/jcb.200801096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Toth J.I., et al. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12873–12878. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mallampalli M.P., Huyer G., Bendale P., Gelb M.H., Michaelis S. Inhibiting farnesylation reverses the nuclear morphology defect in a HeLa cell model for Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14416–14421. doi: 10.1073/pnas.0503712102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Capell B.C., et al. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12879–12884. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Glynn M.W., Glover T.W. Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum. Mol. Genet. 2005;14:2959–2969. doi: 10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- 118.Wang Y., et al. Epidermal expression of the truncated prelamin A causing Hutchinson-Gilford progeria syndrome: effects on keratinocytes, hair and skin. Hum. Mol. Genet. 2008;17:2357–2369. doi: 10.1093/hmg/ddn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kieran M.W., Gordon L., Kleinman M. New approaches to progeria. Pediatrics. 2007;120:834–841. doi: 10.1542/peds.2007-1356. [DOI] [PubMed] [Google Scholar]
- 120.Whyte D.B., et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 121.Yang S.H., Andres D.A., Spielmann H.P., Young S.G., Fong L.G. Progerin elicits disease phenotypes of progeria in mice whether or not it is farnesylated. J. Clin. Invest. 2008;118:3291–3300. doi: 10.1172/JCI35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rogers M.J. From molds and macrophages to mevalonate: a decade of progress in understanding the molecular mode of action of bisphosphonates. Calcif. Tissue Int. 2004;75:451–461. doi: 10.1007/s00223-004-0024-1. [DOI] [PubMed] [Google Scholar]
- 123.Scaffidi P., Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang S., et al. Correction of cellular phenotypes of Hutchinson-Gilford Progeria cells by RNA interference. Hum. Genet. 2005;118:444–450. doi: 10.1007/s00439-005-0051-7. [DOI] [PubMed] [Google Scholar]
- 125.Fong L.G., et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J. Clin. Invest. 2006;116:743–752. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wolf C.M., et al. Lamin A/C haploinsufficiency causes dilated cardiomyopathy and apoptosis-triggered cardiac conduction system disease. J. Mol. Cell. Cardiol. 2008;44:293–303. doi: 10.1016/j.yjmcc.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Arimura T., et al. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. . Hum. Mol. Genet. 2005;14:155–169. doi: 10.1093/hmg/ddi017. [DOI] [PubMed] [Google Scholar]