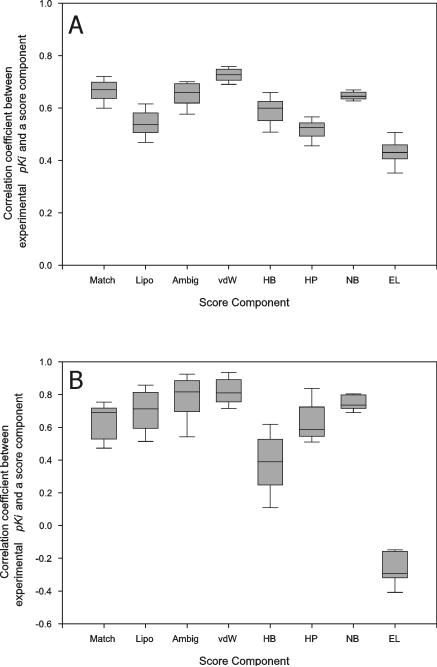
Figure 5.
The distribution of the correlation coefficients between score components and experimental pKi's. The ligands of CDS7 (A) and CDS6b (B) were cross-docked to the receptor structures of CDS7 and CDS6b, respectively, with FlexX (for Match, Lipo, Ambig, vdW, HB and HP columns) or AutoDock (for NB and EL columns) and their docked conformations ranked with FlexX (for Match, Lipo and Ambig columns), X-Score (for vdW, HB and HP columns) or AutoDock (for NB and EL columns) as described in Material and Methods. The best scoring conformations of the ligands for each receptor structure were collected and the correlation coefficient between score components and experimental pKi was calculated for each receptor and its best-scoring ligand conformations. The correlation coefficients from all the receptor structures were pooled to obtain the shown distribution. The binding score components meant the following according to the developers6,8,34; Match: ionic, hydrogen bond and aromatic interaction score of FlexX, Lipo: lipophilic contact score of FlexX, Ambig: hydrophilic-lipophilic contact score of FlexX, vdW: van der Waals interaction score of X-Score, HB: hydrogen bond score of X-Score, HP: hydrophobic interaction score of X-Score, NB: van der Waals interaction and hydrogen bond score of AutoDock and EL: electrostatic interaction score of AutoDock.