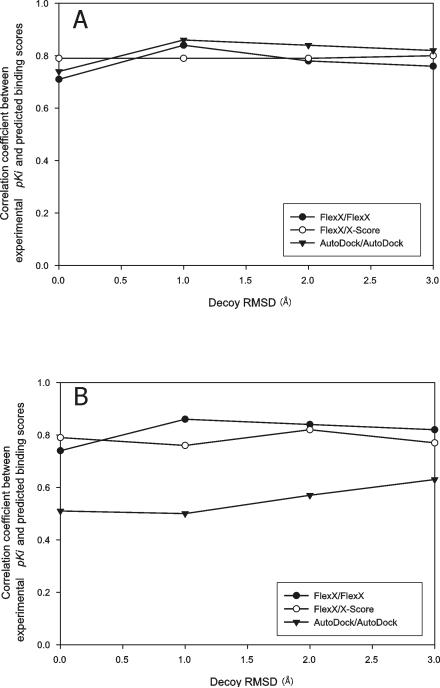
Figure 6.
Correlation between experimental pKi and predicted binding score from decoy receptor structure docking. The ligands of CDS7 (A) and CDS6b (B) were docked to the deformed decoys of the receptor structures of the complex structures 1oyq of CDS7 and 1tlp of CDS6b, respectively. For each ligand, the “best-of-best scoring” complex was chosen in each decoy Cα RMSD from native bin and the binding affinity estimate from this complex was used to calculate CC. The legend indicates docking/ranking programs used. Decoy RMSD of 0 Å means native receptor structures.