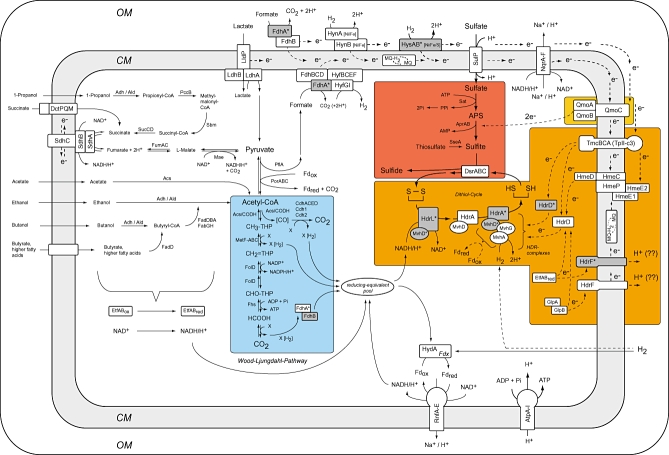
Fig. 1.
Metabolic reconstruction of Db. autotrophicum HRM2 based on known growth substrates and metabolic capacities. Complete oxidation of organic substrates and CO2 fixation under autotrophic growth conditions proceeds via the Wood–Ljungdahl pathway (blue box). The sulfate reduction is given in a dark red box, heterodisulfide reduction is given in a dark orange box and electron transfer from Qmo to sulfate reduction is given in an orange box. All selenium-dependent proteins are marked by dark grey backgrounds and an asterisk (*). The cytoplasmic membrane is given in light grey. Arrows indicate metabolic flows; dashed lines indicate assumed or putative electron flows. For reasons of simplicity, cytochromes, multihaem proteins and the following proteins with one or more paralogues are displayed only once in the figure: Acs, CysAW, EtfAB, FdhAB, GlpAB, HdrA, HdrL, MvhD, PorABC, RnfA–E, Sat, Suc, SulP, TmcBCA. Abbreviations are: CM, cytoplasmic membrane; OM, outer membrane; Acs/CODH, acetyl-CoA synthase/CO dehydrogenase; Acs, acetyl-CoA synthetase; AcsF, CODH maturation factor; Adh, aldehyde dehydrogenase; AMP/ADP/ATP, adenosine-monophosphate/-diphosphate/-triphosphate; AprAB, adenylylsulfate reductase; AtpA–I, ATP synthase F0/F1; Cit, citrate synthase; CO2, carbon dioxide; Cdh1/2, carbon monoxide dehydrogenase; CdhA–E, bifunctional acetyl-CoA synthetase/carbon monoxide dehydrogenase; DctPQM, TRAP-type C4-dicarboxylate transporter; DsrABC, dissimilatory sulfite reductase complex; EtfABox/EtfABred, electron transfer flavoprotein, oxidized and reduced form; FabGH, 3-oxoacyl-acyl carrier synthase/reductase; FadDBA, long-chain fatty acid-CoA ligase; FdhABCD, formate dehydrogenase; Fdox/Fdred, ferredoxin, oxidized and reduced form; FdrABC, fumarate reductase; Fhs, formate-tetrahydrofolate ligase; Fdx, ferredoxin, 4Fe-4S cluster; Fhs, formate-tetrahydrofolate ligase; FolD, methylenetetrahydrofolate dehydrogenase; FumAC, fumarate hydratase; GlpAB, glycerol-3-phosphate dehydrogenase; HdrADFL, heterodisulfide reductase; HmeCDEP, Hdr-like menaquinol-oxidizing complex; HydA, [Fe]-only fusion hydrogenase; HyfBCDEFGI, hydrogenase Hyf homologue; HynAB, periplasmic [Ni/Fe] hydrogenase; HysAB, periplasmic [Ni/Fe/Se] hydrogenase; Idh, isocitrate dehydrogenase; LdhAB, lactate dehydrogenase; LldP, l-lactate permease; Mae, malic enzyme; MetF-ABC, methylenetetrahydrofolate reductase; MQ/MQ-H2, menaquinone pool, oxidized and reduced form; MvhADG, methylviologen non-reducing hydrogenase; NAD+, NADH/H+, nicotinamide-adenine dinucleotide, oxidized and reduced form; NADP+, NADPH/H+, nicotinamide-adenine dinucleotide phosphate, oxidized and reduced form; NqrA–F, Na+-translocating NADH-quinone reductase; PccB, propionyl-CoA carboxylase; Pfl, pyruvate formate lyase; PorABC, pyruvate:ferredoxin oxidoreductase; PPi, pyrophosphate; QmoABC, quinone-interacting membrane-bound oxidoreductase complex; RnfA–E, electron transport complex protein; Sat, sulfate adenylyl transferase; Sbm, methylmalonyl-CoA mutase; SdhABC, succinate dehydrogenase/fumarate reductase; SseA, thiosulfate sulfur transferase; SucCD, succinyl-CoA synthetase; SulP, high-affinity H+/SO42− symporter; TmcABC, acidic type II cytochrome complex; Tst, thiosulfate sulfur transferase; X/XH2, unknown carrier of reducing equivalent, oxidized and reduced form.