Figure 2.
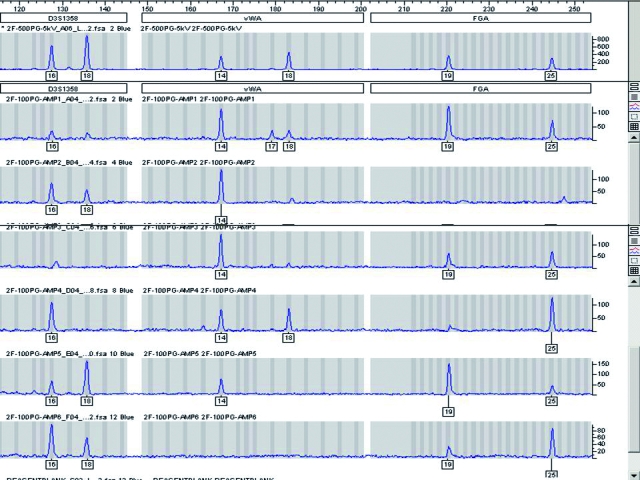
A known DNA sample divided into 6 reactions, each containing 100 pg of DNA and independently amplified using the AmpFlSTR® Profiler Plus® ID PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA) and following the manufacturer’s recommended amplification conditions including 28 cycles for the polymerase chain reaction, but using a 40 relative fluorescence unit (RFU) detection threshold. The example shows the results for a subset of the full profile (D3S1358, vWA, and FGA loci). The top panel is a control sample run with sufficient DNA for conventional typing. The 6 panels below the control are the 100 pg replicates. This example demonstrates that replicates of pristine control samples containing 100 pg of DNA or less may not yield reproducible results. The problems associated with the amplification and interpretation of low copy number (LCN) samples are greatly compounded when evidentiary mixtures are analyzed. The lack of reproducibility will persist with methods employed to increase sensitivity of detection for LCN typing.
