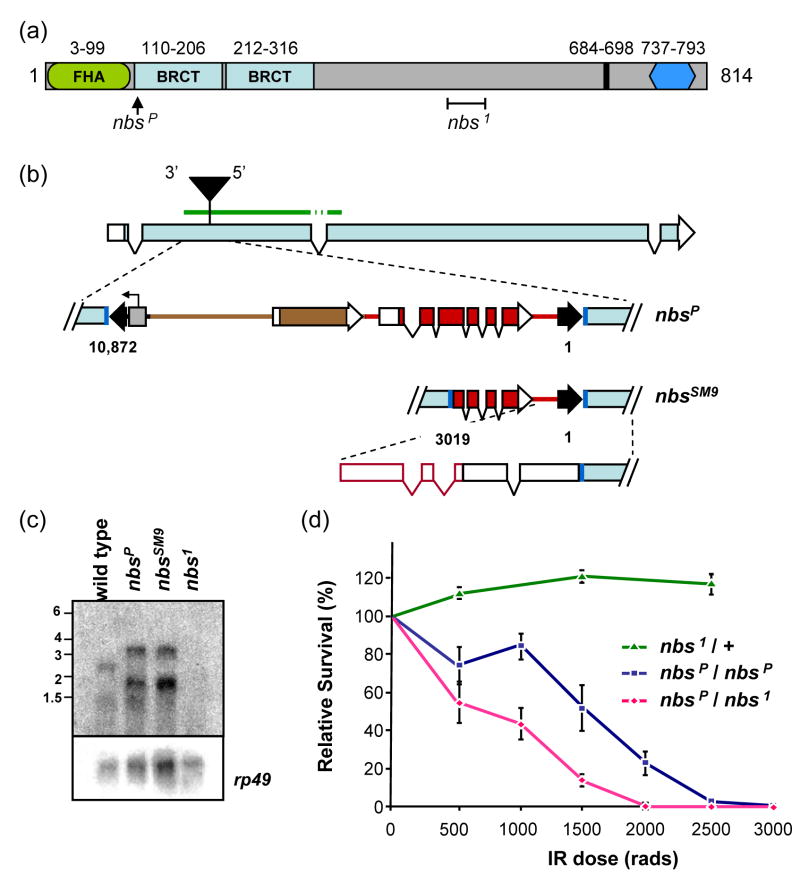
Figure 1. The Drosophila NBS protein and nbs mutations.
(a) The predicted Drosophila melanogaster NBS protein is 814 residues. Conserved sequences include an FHA domain (green oval), two BRCT repeats (light blue rectangles), a putative nuclear localization signal (black line), and the MRE11 binding site (dark blue hexagon). The positions of two mutations are indicated: nbsP (insertion of a P element) and nbs1 (238-bp deletion denoted by a bracket).
(b) Structure of nbsP and nbsSM9 mutations. The nbsP allele is caused by insertion of a 10-kb P element construct into coding sequences in the second exon of nbs. Dark blue lines represent the 8-bp insertion site sequence that is duplicated at each end of the P element. Black arrowheads indicate P element sequences, including inverted repeats at the ends that are required in cis for transposition. This construct carries a y+ body color gene and a w+ eye color gene (brown and red, respectively; rectangles indicate exons; protein-coding sequences filled), and a promoter under the control of an S. cerevisiae UAS sequence (gray box; arrow indicates start and direction of transcription). The P element is inserted such that the 5′ P element end is near the 3′ end of nbs. The nbsSM9 derivative has lost approximately 7 kb, including one inverted repeat, all of y+, and much of the w+ gene, retaining nucleotides 1-3019. The diagram at the bottom of this panel depicts the structure of the transcript determined by 5′ RACE. The green bar above the illustration of the intron/exon structure indicates the position of the probe used to detect transcripts (the probe was from cDNA, so intron and P element sequences were not included).
(c) Transcripts in nbs mutants. A blot of polyA-selected RNA from wild-type and nbs mutant L3 larvae was probed with a fragment from the 5′ end of nbs. Transcripts of about 1200 and 2600 nt are detected in wild-type larvae. In nbsP and nbsSM9 mutants, larger transcripts (1900 and 3300 nt) are seen; no transcripts are detected in the nbs1 mutant. The membrane was stripped and re-probed for rp49 as a loading control.
(d) Sensitivity of nbs mutants to ionizing radiation. An IR sensitivity assay of nbs mutants was carried out at the doses of gamma irradiation indicated on the X axis. Each point is the mean survival of mutants relative to survival of heterozygous siblings (for nbsP/nbsP and nbs1/nbsP) or wild-type siblings (for nbs1/+). Error bars indicate standard deviation (n = 6–14 bottles for each point).