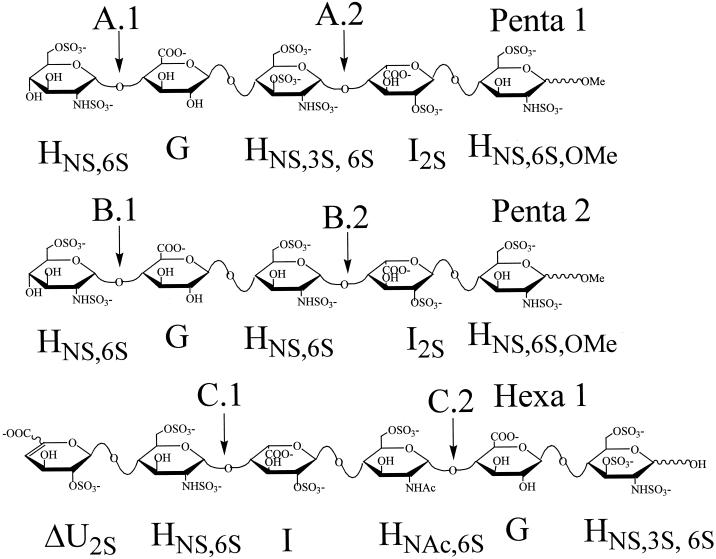
Figure 1.
Structures of the three oligosaccharide model compounds used in this study. (A) Pentasaccharide 1 (Penta 1) has the sequence HNS,6SGHNS,3S,6SI2SHNS,6S,OMe, contains a fully intact AT-III binding site, and has a calculated molecular mass of 1508.2. The two glycosidic linkages potentially susceptible to heparinase I, II, or III cleavage are labeled A.1 and A.2. (B) Pentasaccharide 2 (Penta 2), with the sequence HNS,6SGHNS,6SI2SHNS,6S,OMe and a calculated molecular mass of 1428.1, is structurally identical to Penta 1, less a 3-O sulfate on the internal glucosamine, thus it does not contain a full AT-III site. As with Penta 1 the bonds potentially susceptible to heparinase cleavage are marked B.1 and B.2. (C) A heparinase-derived hexasaccharide (Hexa 1), with the sequence ΔU2SHNS,6SIHNAc,6S,GHNS,3S,6S, also was used in this study. Hexa 1 (calculated molecular mass 1614.3) contains only a partially intact AT-III binding site; similar to AT-10 it is missing the reducing end I2SHNS,6S disaccharide unit. As with Penta 1 and Penta 2, sites of potential cleavage are marked C.1 and C.2.