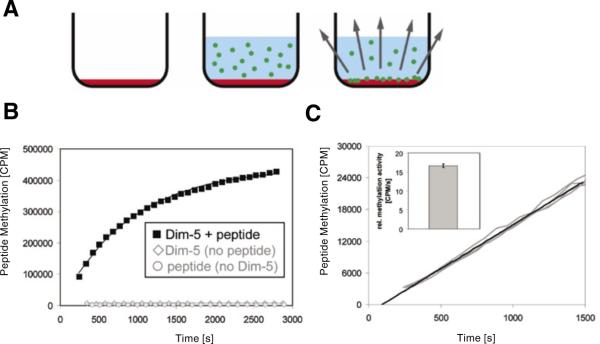
Figure 1. Experimental design.
(A) Drawing of the principle of the continuous peptide methylation assay using FlashPlates with scintillator embedded into the walls of the microplate. In the first step, the well of a FlashPlate is coated with target peptide (red shading). Then, enzyme and radioactive coenzyme are added (radioactively labeled methyl groups are depicted by green circles). The transfer of the methyl groups to the target peptide leads to a close approximation of radioactive methyl group and scintillator, which results in a scintillation signal. (B) After adding radioactive S-adenosyl-L-methionine (AdoMet) and enzyme (3.5 μM) to the well of a FlashPlate coated with 320 pmol peptide, a strong scintillation signal appeared that reflects the progress of the methylation reaction. If enzyme is omitted or if the plates are not coated with peptide, no signal change was detected. (C) Reproducibility of the assay. Four independent experiments using 3.5 nM enzyme were carried out in different wells coated with 320 pmol peptide, and the results were overlaid without any data normalization. Initial methylation rates were determined by averaging data points over the first 1500 s. As shown in the insert, the initial methylation rates determined by the slope were identical within ±3% error.