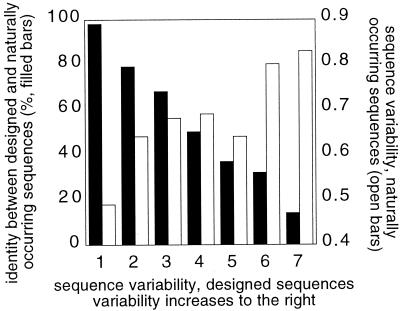
Figure 3.
Sequence conservation in designed sequences correlates with sequence identity to the native sequence and sequence conservation in protein families. When the design program shows a strong preference for a particular amino acid at a sequence position, it more often prefers the native amino acid, and the residue is likely to have low sequence variability in naturally occurring sequences. Each position in each redesigned protein was assigned to a bin (x axis) based on the sequence entropy (∑frequency(aai)⋅ln(frequency(aai)) summed over all 20 amino acids, aai) at the position in a large set of sequences generated by the Monte Carlo search procedure (the numbers of residues in bins 1–7 are, respectively, 86, 91, 91, 126, 107, 79, and 94; higher sequence entropy is to the right). The left y axis indicates the percentage of residue positions that had the native amino acid in the designed sequences. The right y axis indicates the average sequence variability observed in naturally occurring sequences as derived from multiple sequence alignments (MSAs). The MSAs were taken from HSSP files (21). Results are shown for core residues. Only residue positions that had at least 10 sequences in the MSA were used (60 proteins total).