Abstract
The dependence of polyketide synthase and terpene cyclase mechanistic adaptation on the chemistry of their oligomeric substrates illuminates a convergent evolutionary strategy for shaping cyclization in these otherwise disparate reactions. Evolution of these enzyme families relies on rhythmic tangos, in which the enzymes and substrates together determine product outcome by negotiating decision networks governing intrinsic and induced chemical reactivities.
The numerous and varied chemical ecologies of the natural world rely on the emergence and ongoing evolution of secondary metabolism, as species fortuitously discover and adaptively refine biosyntheses of unique collections of natural products. These specialized metabolites allow their hosts to communicate with their environments and flourish in particular niches. Generally speaking, a comprehensive metabolic tree encompassing both primary and secondary metabolism helps define each species. Catabolism (the ‘roots’ of the tree) liberates energy stored in molecules while dismantling them into simpler and recyclable building blocks (the tree ‘trunk’) for reallocation into various biosynthetic pathways (the tree ‘limbs’) where chemical diversity is rebuilt. Specialized metabolites arise via species-specific evolutionary divergence from primary metabolism. More often than not, it is these branches defining secondary metabolism that contribute in notable ways to the individuality of each species.
Within these secondary metabolic branches, a myriad of cyclic terpene and polyketide molecules form the core of much specialized chemical complexity. Arising through differing intramolecular condensations of a smaller collection of oligomeric substrates, in turn created by repetitive linear condensations of acetyl or isoprenoid building blocks (Fig. 1), these cyclic secondary metabolites are biosynthesized by a few ‘uber-adaptable’ enzyme superfamilies originating in primary metabolism. Recent discussions of enzyme evolution1 have focused on these remarkable enzymes because of their richly branching metabolic variety, unusual product promiscuity and facile functional divergence.
Figure 1.
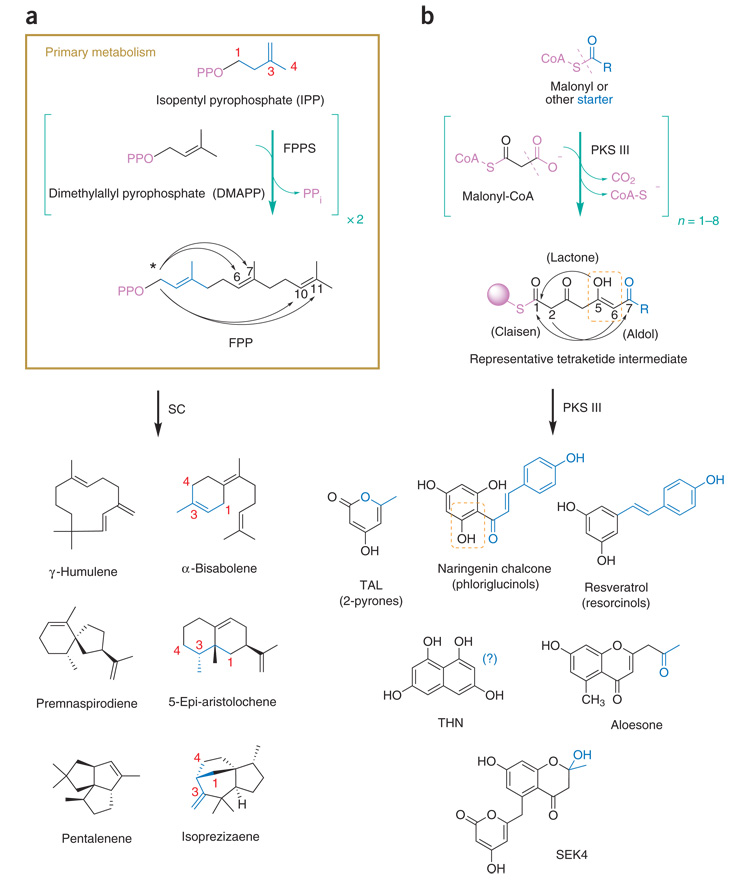
Metabolic diversity via divergent intramolecular cyclization. Building blocks, linear oligomeric intermediates and representative uni-, bi- and tricyclic products biosynthesized by two cyclase families are shown. Leaving groups (violet) are exploited for enzyme recognition and reactivity enhancement. Curved arrows indicate alternate initial intramolecular condensations. (a) SCs cyclize FPP, an isoprenoid trimer from primary metabolism, to biosynthesize hundreds of distinct cyclic sesquiterpene hydrocarbons. Some initial intramolecular condensations of FPP-derived carbocations require a prior isomerization event to a cisoid farnesyl cation (arrows marked by *), whereas others proceed by direct attack of the nascent carbocation following ionization. A highlighted isoprene unit (blue) reveals additional rearrangements en route to SC products (see text). (b) PKS IIIs selectively transfer a CoA-thioester–activated starter moiety (blue) to a catalytic cysteine, catalyze iterative polyketide chain extension via decarboxylative condensations with malonyl-CoA and then divergently cyclize the resulting linear oligomeric intermediates to offload products. Subunits of extended polyketide intermediates can undergo keto-enol tautomerization (dashed orange box). Note that the internal symmetry of the THN product obscures the final position of the incorporated starter moiety (blue).
Structure-guided studies have attempted to elucidate specific enzymatic principles underlying polyketide and polyisoprenoid cyclization mechanisms. Our own efforts focus on two distinct and architecturally simple model systems: the sesquiterpene cyclase (SC)2 and type III polyketide synthase (PKS III)3 families. These enzyme families respectively catalyze the divergent cyclization of linear isoprenoid trimers (Fig. 1a) and variably elongated polyketides (Fig. 1b). As we describe here, juxtaposition of these structurally unrelated ‘cyclase’ (generic SC or PKS III) families reveals a convergent recipe for mechanistic plasticity (Fig. 2). Integral to this general model for mechanistic adaptability is the complementary interplay between the cyclases’ catalytic properties and the intrinsic or induced chemical traits of their cognate substrates.
Figure 2.
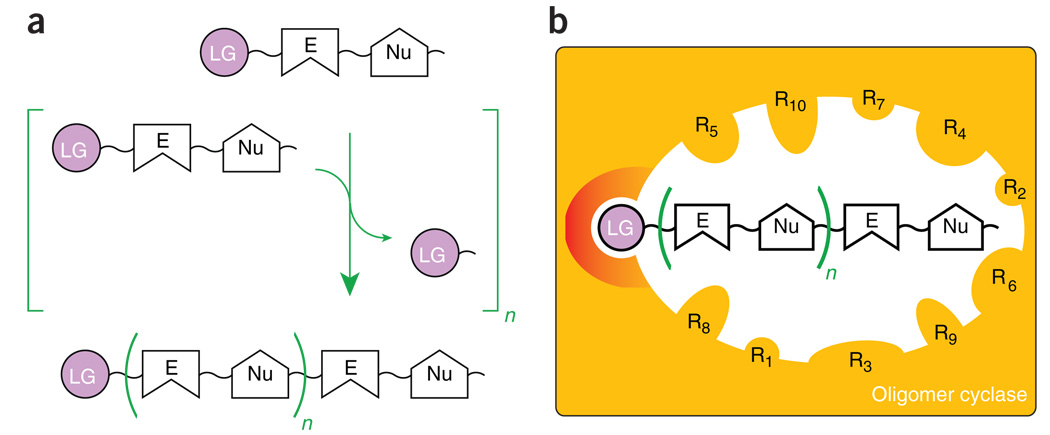
Convergent features of ‘uber-adaptable’ cyclases and their oligomeric substrates. (a) Polymerization involves covalent bond formation between a nucleophile (Nu) and an electrophile (E). Enzymes exploit high-energy bonds tethered to favorable leaving groups (LG) to activate adjacent electrophiles. These moieties also provide convenient chemical handles for modular small-molecule recognition. (b) A generic cyclase is depicted as an orange box. Structural analyses of unrelated PKS IIIs and SCs reveal analogous internal active site cavities lined by amino acid side chains (R), allowing evolving steric control over oligomeric substrate/intermediate conformations leading to alternative juxtapositions of the repetitive and reactive Nu-E pairs. Evolutionary conservation of catalytic machinery (red) for binding and manipulating LGs is also shown.
Chemical intuition and enzymatic studies suggest that SCs and PKS IIIs convergently exploit inherently redundant chemical properties of their respective polyisoprenoid and polyketide substrates/intermediates during the process of natural selection. Thus, mechanistic interpretations of cyclase structure-function experiments that focus on product specificity or promiscuity require a complementary consideration of the intrinsic or induced chemistry of substrates and intermediates before a general model for cyclase evolution can be articulated. This unifying ‘chemical biology’ perspective reveals an intimate connection between the extensively branched metabolic diversity arising from each of these enzyme families and corresponding mechanistic decision networks negotiated within each cyclase active site. These nested network architectures (branching and tree-like) underlie the dynamic partitioning of reaction intermediates. As these reactive intermediates navigate along diverging routes, their alternative fates depend on choreographed negotiations between enzyme and oligomeric intermediates. It is useful to view each multistep reaction pathway as an intricate tango, in which the enzyme and small-molecule dance partners each play critical roles in determining the final product (Fig. 3). Interestingly, comparison of SCs with PKS IIIs in light of this convergent recipe suggests that distinctions in their particular mechanistic plasticity arise from intrinsic differences in their respective small-molecule dance partners.
Figure 3.
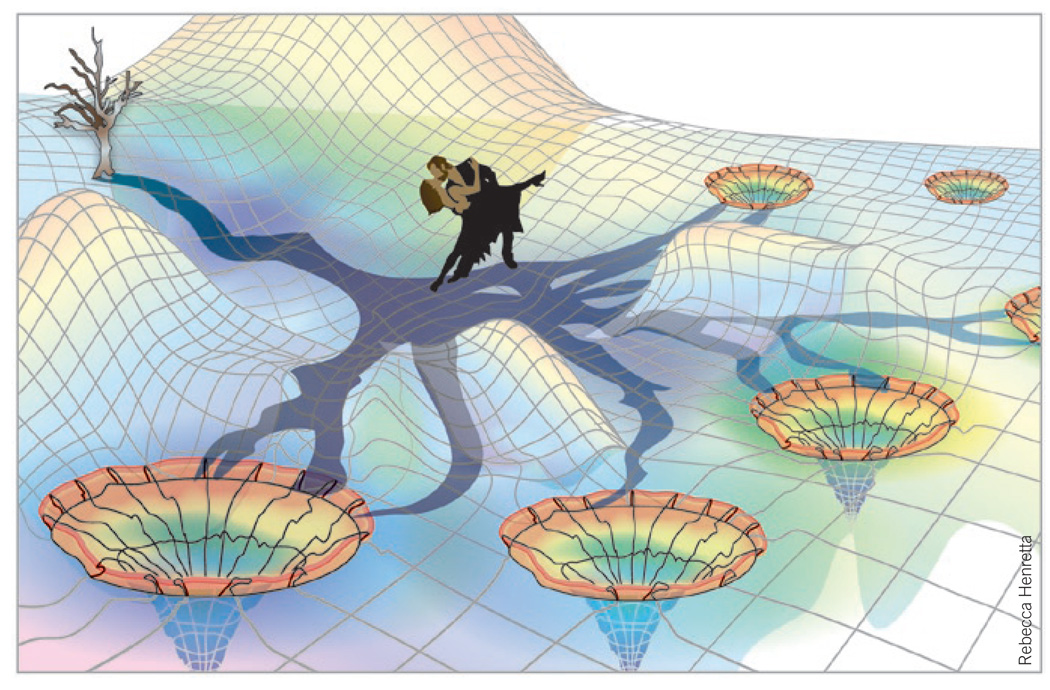
Biosynthetic tangos in the shade of a branching metabolic diversification tree. The vertical tree represents the accessible product spectrum of a particular cyclase-substrate dance pair and is rooted at the edge of the (enzyme-bound) small-molecule conformational energy landscape. The tree casts its shadow across this uneven dance floor, thus highlighting a corresponding horizontal mechanistic decision network. Multistep negotiation of this network across the dance floor constitutes the ‘biosynthetic tango’ between substrate and enzyme that determines the accessibility of alternative reaction pathways leading to distinct stable products.
Emergence of cyclases from primary metabolism
SCs use farnesyl pyrophosphate (FPP) as a substrate (Fig. 1a). FPP turnover initiates upon pyrophosphate loss (ionization), and the resultant carbocation partitions down one of several intramolecular condensation pathways. Further SC-catalyzed reaction partitioning is achieved by intramolecular alkyl and hydride migrations and carbocation neutralization through proton loss or water capture. In some cases, reformation of carbocations from neutral intermediates ensues, followed by additional rounds of alkyl and hydride migrations en route to a neutral hydrocarbon or alcoholic product through proton loss or water capture, respectively. These compounds include hundreds of distinct a-, uni-, bi- and tricyclic hydrocarbons, with rings ranging in size from 3 to 11 carbons, many having multiple chiral centers (Fig. 1a). In contrast, PKS IIIs select a coenzyme A (CoA)-activated starter moiety, and then catalyze between 1 and 8 cycles of iterative acetyl chain extension via decarboxylative condensation reactions. Finally, disparate intra-molecular condensation mechanisms off-load substituted uni-, bi- and tricyclic hydroxylated polyketides (Fig. 1b). Less structurally diverse than terpenes, all PKS III rings are hexacyclic, and most represent substituted phloriglucinols, resorcinols or 2-pyrones (Fig. 1b). Elimination of polyketide hydroxyls4 or hydroxylation of aliphatic terpenes by water capture of carbocation intermediates5,6 can somewhat temper these differences (Fig. 1).
In the late 1990s, the first crystal structures of a PKS III and a plant SC—alfalfa chalcone synthase (CHS)7 and tobacco 5-epiaristolochene synthase (TEAS)8, respectively—appeared, thus opening the door for subsequent structure-guided analyses of functional divergence within each cyclase family. These initial structures also revealed important architectural and mechanistic vestiges of their evolutionary emergence from separate and ancient condensing enzymes of primary metabolism. Notably, each cyclase family (PKS III and SC) conserves primordial three-dimensional structures and key catalytic residues with their extant cousins in primary metabolism that also use recognition of substrate ‘handles’ and subsequent exploitation of kinetically stable yet thermodynamically labile high-energy bonds (Fig. 1 and Fig. 2). While these inferences are specific to SCs and PKS IIIs, the structural and mechanistic links with evolutionarily restrained enzymes from primary metabolism represent widely applicable principles governing the emergence, ancestry and product expansion of secondary metabolism from slowly varying primary metabolism. In fact, the rapid product diversification of secondary metabolic enzymes within the constraints of their structural and mechanistic conservation with primary metabolic enzymes provides the first clue for a common recipe underlying cyclase catalytic emergence and promiscuity.
For polyisoprenoid metabolism, the conserved DDXXD motif in TEAS and other SCs coordinates divalent metal cations to bind and catalytically cleave the pyrophosphate moiety from FPP. This divalent cation recognition motif derives from ancestral isoprenoid diphosphate synthases where, in contrast to SCs, divalent cations are used to catalyze intermolecular isoprenoid condensation to produce linear polyisoprenoid diphosphates like FPP (Fig. 1a). This Mg2+-dependent isoprene activation motif, conserved within the same α-helical protein fold shared between primary and secondary metabolism, implies a relatively facile evolutionary conversion from primary, uniform and ubiquitous intermolecular linear condensations (polymerization) of five-carbon isoprene units to specialized and varied intramolecular condensations (cyclization) of a productively folded polyisoprenoid chain.
Likewise, the homodimeric CHS structure revealed a Cys-His-Asn catalytic triad accessed through a narrow CoA-binding tunnel7. The conserved condensing machinery and homodimeric αβαβα-fold architecture are nearly identical to that of the primary metabolic enzyme β-ketoacyl synthase (KAS) III, which non-iteratively catalyzes the first acetyl extension step of plant and bacterial fatty acid biosynthesis (FAS). Both enzymes use this conserved catalytic triad to load a CoA-activated starter moiety onto the active site cysteine, and then biosynthesize a β-keto thioester product via decarboxylative condensation of a malonyl unit (Fig. 1b)3. In contrast to the evolutionary emergence of terpene cyclases, where intrasimply replaces intermolecular condensation, the emergence of PKS III cyclization is evolutionarily linked to the development of iterative polyketide biosynthesis, which may have been triggered by an active site mutation causing reorientation of the loaded KAS III starter3.
A common recipe for cyclase catalytic promiscuity
Most primary and secondary metabolic enzymes contributing to the overarching metabolic tree of each species tend to recognize their substrates using a chemical ‘handle’. These handles can be an integrated functionality such as a hydroxyl group or a prosthetic attachment such as pyrophosphate or CoA groups. Functional divergence of such enzymes typically involves simple substitution of different but similarly tagged substrates, in an otherwise mechanistically conserved reaction. This trend results in a somewhat scattered distribution (often modulated by temporal or spatial compartmentalization) of most biosynthetic enzyme families across the overall metabolic tree of a species. The evolutionarily conserved mechanistic contribution of the labile chemical handles recognized by the ‘oligomer cyclase’ families will be discussed again shortly (Fig. 1 and Fig. 2). Though compartmentalization and substrate modulation also remain important physiological controls, the most impressive and unusual feature of these cyclases is their ability to catalyze extensive metabolic diversification of conserved substrates. This richly branched expansion mimics the upper biosynthetic portion of the metabolic trees they ultimately serve. Moreover, the mechanistic decision networks underlying these cyclases hint at their special ability for achieving impressive reaction divergence: exploitation of the spatially distinct yet chemically redundant reactivities of their small-molecule substrates.
Through static surfaces or dynamic conformational changes, both SC and PKS III families afford extensive steric control of substrate conformation and solvent accessibility (Fig. 2). Prior to available structural information for either enzyme family, chemical logic suggested that biosynthesis of alternatively cyclized products from shared linear intermediates necessitated juxtaposition of different substrate atoms. This steric collocation of chemically reactive atoms implies the selective sequestration of discrete and productive conformations of substrates and intermediates modulated sterically by the static and/or dynamic active site contours and reactivities. For example, Cane’s formulation of a general stereochemical framework of sesquiterpene biogenesis elegantly delineates a steric modulation model shaping these productive conformations of FPP whereby individual double bonds of the substrate are mutually perpendicular to a common plane9. These stacked arrangements of the double bond pi systems provide a cogent and simple geometric blueprint to explain the myriad of electrophilically driven transformations in sesquiterpene biogenesis.
A second PKS III crystal structure strongly supported this steric modulation model, and lead to the facile conversion of CHS activity to that of a 2-pyrone synthase (2PS) via constriction of active site volume by mutation of only 3 out of ~100 residue differences10 (TAL; Fig. 1b). Likewise, Yoshikuni et al.11 achieved a similar modulation of SC product specificity by structure-guided saturation mutagenesis of the active site cavity in the naturally promiscuous γ-humulene synthase.
Given that evolution does not contemplate the location of effective mutations, it is not surprising that substitutions within second- or third- tier layers surrounding the active site cavity can also adjust reactivity. This notion was demonstrated by comparative structure-function studies of TEAS and its functionally divergent relative henbane premnaspirodiene synthase (HPS), leading to the identification of accumulated substitutions that effectively transmute their respective activities12. In similar fashion, the repeated PKS III evolutionary transition from CHS to stilbene synthase (STS) also involves different sets of buried mutations13. What emerges from both examples is a deeper appreciation for reactivity control shaped by sequence variation beyond the active site surface.
Likewise, an indication that steric modulation alone is insufficient for controlling or inducing reactivity in substrates and intermediates emerged from two separate PKS III mutational studies13,14, which together led to a more comprehensive model called reaction partitioning15. First, an isosteric mutation (noncatalytic triad cysteine to serine) in the active site of 1,3,6,8-tetrahydroxynaphthalene synthase (THNS) redirects THN biosynthesis toward the lactonized triketide TAL previously observed in 2PS (Fig. 1b)14. Whereas steric modulation limits starter selection, polyketide length and cyclization in 2PS (ref. 10), the roomy THNS active site cavity must actively prevent premature triketide cyclization in order to catalyze the synthesis of the pentaketide intermediate preceding THN offloading14. Second, in the case of the repeated evolutionary emergence of STSs from CHSs, structural, mutagenic and mechanistic studies demonstrated an emergent thioesterase-like hydrogen bonding network in STSs caused by subtle repositioning via distal mutations of a conserved active site threonine to redirect the cyclization of the same tetraketide intermediate used by CHS (ref. 13; Fig. 1b).
Interestingly, most polyisoprenoid and polyketide cyclases produce multiple minor products derived from common intermediates. Such ‘derailments’ are perhaps not surprising for multifunctional PKS III enzymes, which must juggle iterative polymerization and reaction terminating cyclization in the same active site. However, this trend toward product promiscuity is mirrored in the more specialized SCs. Students of evolution will recognize this tendency toward multiple products as a valuable precursor to adaptive change, since any desired activity must be present at trace levels before natural selection can reinforce and refine it. Basal persistence of multiple ‘traits’ (mechanistic paths) clearly increases the ‘population pool’ (products) from which natural selection can choose. Moreover, analysis of the source of these persistent alternative paths illuminates the contribution of the small molecule in determining product specificity or promiscuity.
The enzyme-substrate biosynthetic tango
The catalytic promiscuity associated with these cyclase families depends upon shared characteristics of their respective substrates and reaction intermediates. We have discussed the first of these in relation to cyclase evolution from primary metabolism, namely the retention of chemical ‘handles’ such as activating thio- or phosphate-ester bonds to CoA or inorganic pyrophosphate, respectively (Fig. 1 and Fig. 2). By orienting bound substrates and lowering initial activation energies through co-participation with the enzyme dance partner, the resulting leaving groups facilitate reaction triggering. However, the true mechanistic ‘secret weapon’ of these evolutionarily versatile enzyme families is exploitation of repetitive electrophilic and nucleophilic functionalities in each oligomeric substrate (Fig. 1 and Fig. 2). Coupled with sufficient conformational flexibility for enzyme-mediated juxtaposition of complementary pairs of these functionalities, this intrinsic and repetitive reactivity can be easily tuned by natural selection to favor alternative sesquiterpene or polyketide products.
An uneven energy landscape determines the accessible conformations of any flexible substrate in solution. The various conformations and reactivities available to chemically repetitive polyketides or polyisoprenoids result in a more complex energy landscape. Binding within a statically and/or dynamically restrained enzyme active site causes ‘tectonic shifts’ in this landscape that drastically remodel its topography to produce the conformation-dependent reaction coordinates of the enzyme-catalyzed multistep reaction.
Each enzyme-dependent intermediate or product corresponds to an accessible shallow or deep energy well distributed across this varying landscape (Fig. 3). Ingrained in the oligomeric intermediates of our cyclases are a multitude of relatively isoenergetic cyclization fates. Distinct energetically favorable routes connecting a given substrate or intermediate to alternative products enforce diverging reaction paths available to each dance pair. These paths comprise the spatial and temporal mechanistic decision networks co-negotiated by the dance partners. Collectively, these decisions determine each cyclase’s richly branching contribution to the much larger overall metabolic diversification tree of their host organism.
It is useful to imagine just the cyclase-relevant branching portion of the overarching metabolic tree as a smaller vertical tree rooted at the edge of the landscape and casting its shadow across the uneven dance floor. This shadow corresponds to decision networks previously described and highlights only energetically accessible reaction paths (Fig. 3). Indeed, just as real limb shadows often overlap, so can different sets of protein-directed conformational poses during the multistep dance convergently lead to the same final product. Evolution sculpts each malleable enzyme active site around its limber dance partner as they traverse the dance floor together. Natural selection, in turn, choreographs the dance toward an existing or emergent branch of the shadow cast on the convoluted dance floor.
Vive la difference!
Intrinsic reactivity differences between linear polyketides and polyisoprenoids illuminate the first major difference between PKS III and SC biosynthetic strategies: the modular separation of enzymes catalyzing isoprenoid diphosphate polymerization (substrate biosynthesis) and subsequent cyclization. In contrast, polymerization and cyclization are coupled in nonreducing PKS systems, and the dearth of extended and unreduced linear polyketides in nature hints at their intrinsic instability. Conversely, there is a plenitude of stable polyisoprenoids in nature, from geranyl (C10) to decaprenyl (C50) diphosphates16, including carotenoids17 and natural rubbers composed of a thousand or more isoprene units.
While our recipe for cyclase uber-adaptability demands a degree of flexibility from each linear polymer, a tetraketide intermediate may have either more or fewer degrees of freedom than a comparable di-isoprenoid linear chain, depending on the extent of enolization (Fig. 1). It remains unclear how much subsequent cis or trans enolization of each newly formed β-keto unit occurs during the short lifetime of the enzyme-sequestered linear intermediate. It seems more likely that drastic differences in stability between polyisoprenoids and unreduced polyketides instead arise from the accentuated reactivity of polar two-carbon acetyl units when linked into β-keto polymers.
Indeed, this alternating polarity of polyketides exclusively favors the chemically facile formation of six-member rings. Reaction-terminating six-member ring formation at the tri- or tetraketide stage tends to preclude further iterations of PKS III chain extension, greatly biasing reaction specificity toward single-ring products3. Thus, iterative polyketide chain extension and control of competing intrinsically favorable cyclizations must be kept in balance by PKS IIIs.
In contrast, the relative inertness of FPP allows the SC family to more freely explore substrate conformational space and pose their allylic polymers before the tango ensues. The proper juxtaposition of C1 to either end of the C6,7 or C10,11 nucleophilic double bonds promotes entry into one of four initial cyclization pathways, forming the major branches of the SC diversification tree (Fig. 1a). Once the nascent farnesyl carbocation is formed, however, SCs must likewise tame reactive intermediates. Water capture or proton elimination can effectively quench the reaction in its infancy to produce linear alcohol or hydrocarbon products. Therefore, the SC must stringently control both the timing and location of cation quenching to provide a window of opportunity for various cyclization reactions to occur.
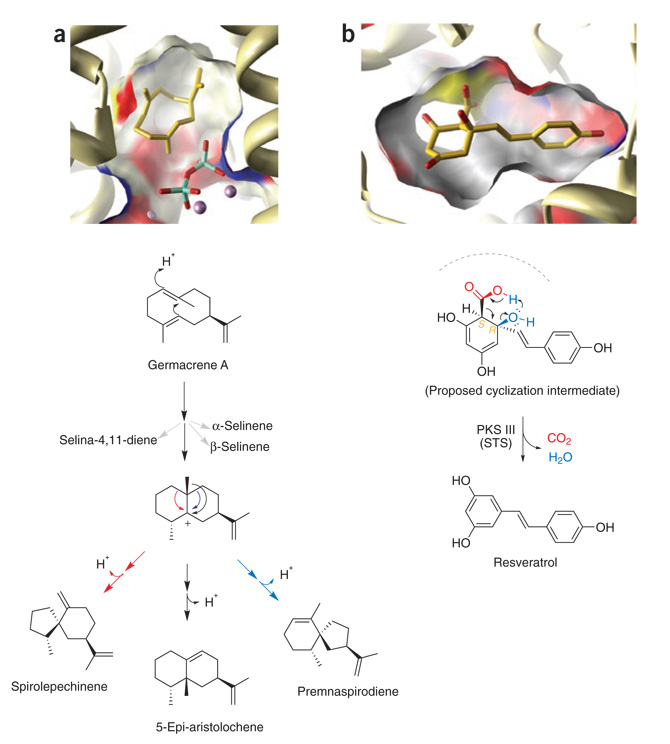
Though acid-catalyzed biomimetic solution-based cyclizations of relatively apolar farnesol or nerolidol preferentially form the six-member bisabolene rings18, the initial four competing SC cyclization pathways nonetheless serve to broaden natural terpene diversity. Notably, six- or seven-member initial ring formation requires prior isomerization to nerolidyl pyrophosphate, providing another step in the overall reaction catalyzed for enzymatic tuning of product specificity. The numerous energetically accessible rearrangements available to allylic carbocations allow much greater reaction divergence downstream of the first cyclization event (Fig. 4a), which is in stark contrast to the rapid and energetically steep aromatic collapse of cyclized polyketide intermediates (Fig. 4b). And unlike aromatic polyketide products, cyclic terpenes often preserve chiral information reflecting the stereochemical course of their multistep reactions (Fig. 1).
Figure 4.
Comparison of postcyclization reactions in the SC (left) and PKS III (right) ‘reaction chambers’. Transient intermediates (gold stick models) are modeled in cut-away surface views of the active site cavities (colored by atom type) and surrounding protein (ribbons). (a) Divergent TEAS-catalyzed cyclization of the neutral germacrene A intermediate. Following Mg2+-assisted elimination of PPi (bottom right of active site figure), TEAS forms germacrene A from the initial C1,10 cyclization and deprotonation of the farnesyl cation (see Figure 1). Protonation of germacrene A regenerates a carbocation to launch a new series of electrophilic cyclizations marked by competing alkyl shifts and terminating in alternative quenching via proton elimination. (b) Rapid collapse to aromaticity obscures the intervening stereochemistry of PKS III cyclization intermediates. Modeled is an unstable and proposed cyclic intermediate of the stilbene synthase (STS)-catalyzed tetraketide aldol condensation leading to resveratrol (see Figure 1), viewed from the perspective of the ‘aldol switch’. Also visible are the catalytic cysteine (yellow surface) and the CoA-binding tunnel. Shown below, the coupled elimination of a carboxylate (red) and adjacent hydroxyl group (blue) following the STS-catalyzed aldol condensation reaction.
In short, the ‘biosynthetic tango’ metaphor emerging from our convergent and generic recipe for mechanistic plasticity also highlights key differences in the intrinsic traits underlying the contrasting trends in cyclized product diversity of SCs and PKS IIIs. In particular, it seems likely that differing biases in product diversity arise almost exclusively from disparate reaction landscapes inherent to the distinct small-molecule dance partners used by SCs and PKS IIIs during cyclization.
A unifying theme for mechanistic adaptation
Fascinating differences aside, SCs and PKS IIIs share a collective recipe for mechanistic adaptability dictated by the evolutionary inter-dependence of enzyme structure and intrinsic/induced reactivities of substrates and catalytic intermediates. Computational models can provide valuable insights into these enzyme-substrate negotiations, as demonstrated by the theoretical treatment of pentalenene biosynthesis, which illuminated multiple energetically feasible cyclization routes leading from the farnesyl cation to pentalenene19. Though informative, current theoretical calculations do not account for correlated motions of enzymes and small molecules. Future advances in theoretical calculations and experimental techniques to quantify correlated motions may enable direct observation of the more subtle intricacies of both cyclase families, whether their biosynthetic dances are rhythmic or eclectic, and whether they are punctuated with dramatic synchronous pauses or consist simply of fluid cascades.
These insights also pertain to polyketide modifications including β-keto reduction4 and incorporation of alternative extender units20, to terpene diversity arising from alternate isoprenoid coupling modes21–23, and (to varying extents) to other equally important polyketide24 and polyisoprenoid2 cyclase families not discussed here. Identification of the key ingredients in this shared recipe for terpene and polyketide cyclization promiscuity, and consideration of the roles of each dance partner in the resulting biosynthetic tango, illuminates the extraordinary evolutionary histories of these uber-adaptive enzyme superfamilies. The realization that facile modulation of product specificity is often an intrinsically substrate-dependent trait of these cyclase families suggests that experiments aimed at exploring reaction plasticity need not start from a particularly promiscuous enzyme. The substrate-dependent product diversity of the SC and PKS III families also cautions against overgeneralization from single examples of observed cyclase functional divergence to all enzymes, or even to all evolution within these respective superfamilies of primary and secondary metabolic enzymes.
However, the restraints that differing small-molecule traits impose on evolutionary divergence of biosynthetic pathways and enzymes constitute a universal theme for mechanistic adaptation and natural selection. While unreduced polyketides are intrinsically unstable, stable growing isoprene polymers can become too large and unwieldy to fold inside a cyclase active site. And unlike terpene cyclases, which can lead by ‘outmaneuvering’ their small-molecule dance partners, iterative PKS III systems find it easier to modulate starter specificity rather than ‘overpower’ the few intrinsically favored cyclization proclivities of their linear intermediates. Modular PKS I systems (reviewed in ref. 24) greatly reduce the intrinsic reactivity of most nascent β-keto moieties, thus lending their linear intermediates a superficial resemblance to polyisoprenoid chains. As elimination of all β-keto moieties during iterative biosynthesis would simply produce an inert fatty acid, these modular PKSs pay a heavy price in protein size to allow ‘custom’ tailored reduction of individual subunits. Coerced by intrinsic polyketide reactivity to adopt this ‘expensive’ strategy, modular PKSs fully capitalize on it for subsequent divergence via continued subunit customization, as opposed to the cyclization complexity that is evolutionarily exploited by less expensive terpene or iterative PKS systems.
ACKNOWLEDGMENTS
The authors acknowledge support from the Howard Hughes Medical Institute, the US National Institutes of Health and the US National Science Foundation.
References
- 1.Fischbach MA, Clardy J. Nat. Chem. Biol. 2007;3:353–355. doi: 10.1038/nchembio0707-353. [DOI] [PubMed] [Google Scholar]
- 2.Christianson DW. Chem. Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 3.Austin MB, Noel JP. Nat. Prod. Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 4.Bomati EK, Austin MB, Bowman ME, Dixon RA, Noel JP. J. Biol. Chem. 2005;280:30496–30503. doi: 10.1074/jbc.M502239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deguerry F, et al. Arch. Biochem. Biophys. 2006;454:123–136. doi: 10.1016/j.abb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Mercke P, Crock J, Croteau R, Brodelius PE. Arch. Biochem. Biophys. 1999;369:213–222. doi: 10.1006/abbi.1999.1358. [DOI] [PubMed] [Google Scholar]
- 7.Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Nat. Struct. Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 8.Starks CM, Back K, Chappell J, Noel JP. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 9.Cane DE. Acc. Chem. Res. 1985;18:220–226. [Google Scholar]
- 10.Jez JM, et al. Chem. Biol. 2000;7:919–930. doi: 10.1016/s1074-5521(00)00041-7. [DOI] [PubMed] [Google Scholar]
- 11.Yoshikuni Y, Ferrin TE, Keasling JD. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 12.Greenhagen BT, O’Maille PE, Noel JP, Chappell J. Proc. Natl. Acad. Sci. USA. 2006;103:9826–9831. doi: 10.1073/pnas.0601605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Austin MB, Bowman ME, Ferrer JL, Schröder J, Noel JP. Chem. Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 14.Austin MB, et al. J. Biol. Chem. 2004;279:45162–45174. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- 15.Austin MB, Noel JP. In: ACS Symposium Series 955, Polyketides: Biosynthesis, Biological Activities, and Genetic Engineering. Rimando AM, Baerson SR, editors. Washington DC: American Chemical Society; 2007. pp. 185–197. [Google Scholar]
- 16.Wang K, Ohnuma S. Trends Biochem. Sci. 1999;24:445–451. doi: 10.1016/s0968-0004(99)01464-4. [DOI] [PubMed] [Google Scholar]
- 17.Umeno D, Tobias AV, Arnold FH. Microbiol. Mol. Biol. Rev. 2005;69:51–78. doi: 10.1128/MMBR.69.1.51-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutsche CD, Maycock JR, Chang CT. Tetrahedron. 1968;24:859–876. [Google Scholar]
- 19.Gutta P, Tantillo DJ. J. Am. Chem. Soc. 2006;128:6172–6179. doi: 10.1021/ja058031n. [DOI] [PubMed] [Google Scholar]
- 20.Song LJ, et al. J. Am. Chem. Soc. 2006;128:14754–14755. doi: 10.1021/ja065247w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang A, et al. Proc. Natl. Acad. Sci. USA. 2004;101:9601–9606. doi: 10.1073/pnas.0401298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunawardena K, Rivera SB, Epstein WW. Phytochemistry. 2002;59:197–203. doi: 10.1016/s0031-9422(01)00438-1. [DOI] [PubMed] [Google Scholar]
- 23.Heathcock CH, Finkelstein BL, Aoki T, Poulter CD. Science. 1985;229:862–864. doi: 10.1126/science.3927485. [DOI] [PubMed] [Google Scholar]
- 24.Staunton J, Weissman KJ. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]