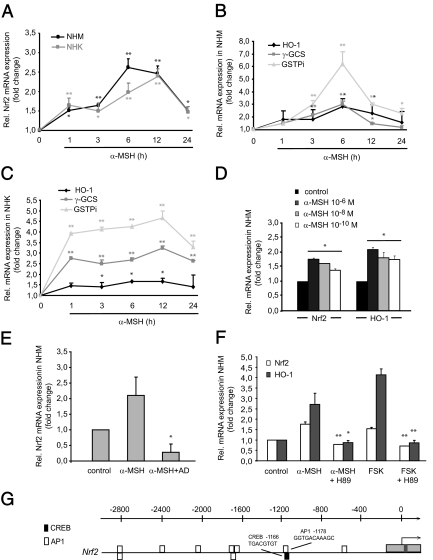
Figure 4.
α-MSH induces Nrf2 and Nrf2-dependent gene expression in vitro and activates the cAMP/PKA pathway. A, Real-time RT-PCR analysis of α-MSH-induced Nrf2 expression in NHM and NHK. Cells were stimulated with α-MSH (10−6 m) for various time points. *, P < 0.01; **, P < 0.001 vs. control. B and C, α-MSH induces mRNA expression of HO-1, γ-GCS, and GSTPi in NHM and HNK as shown by real-time RT-PCR. Cells were stimulated with α-MSH as outlined above. *, P < 0.05; **, P < 0.001 vs. control. D, Dose kinetic analysis of the up-regulating effect of α-MSH on Nrf2 and HO-1 expression in NHM. Cells were stimulated with α-MSH for 6 h as indicated. Nrf2 and HO-1 RNA expression levels were determined by real-time RT-PCR. *, P < 0.001 vs. control. E, Blockade of transcription abrogates α-MSH-induced Nrf2 mRNA expression. NHM were stimulated with α-MSH (10−6 m) for 6 h in the absence or presence of AD (5 μg/ml) followed by real-time RT-PCR analysis of Nrf2. *, P < 0.01 vs. α-MSH. F, Identification of the cAMP/PKA pathway in α-MSH-mediated Nrf2 and HO-1 induction. NHM were treated with α-MSH (10−6 m) or FSK (1 μm) in the presence or absence of the PKA inhibitor H89 (10 μm) for 6 h followed by real-time RT-PCR analysis of Nrf2 and HO-1. *, P < 0.01; **, P < 0.001 vs. α-MSH/FSK. G, Result of the in silico promoter analysis of the promoter regions of Nrf2 for putative CREB and AP1 binding sites. Each block identified by the TOUCAN MotifLocator represents a possible binding site. Black bars, CREB sites; white bars, AP1 sites. The numbers on the axis refer to bases upstream from the transcription start site.