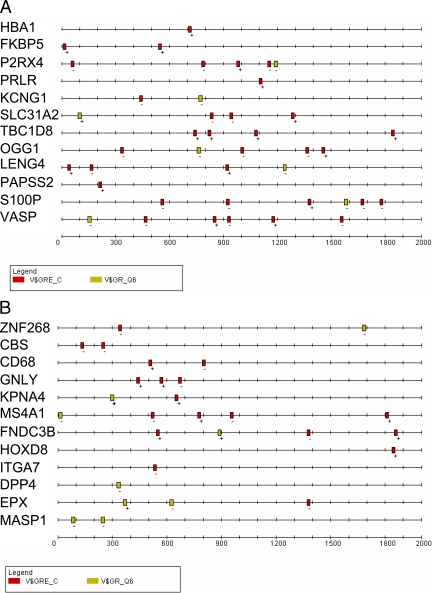
Figure 1.
Position and spacing of putative PRE in 2000-nt proximal promoters of PR-up-regulated or random genes. A, 12 PR up-regulated (20,21,22) and B) 12 random genes were searched for 2000-nt promoter sequences using the EZ retrieve database (23). SRMS was used to define 16-nt conserved motifs and patterns within them. The MEME database was used for large-scale sequence alignment (27). The TRANSFAC database was specifically searched for consensus PRE using two matrices: V$GRE_C (red boxes) and V$GR_Q6 (yellow boxes). Note: the nucleotide numbering on the abscissa is the reverse of conventional promoter positions due to the default numbering in SRMS with 2000 at the putative transcription start site, and ″0″ designates -2000.