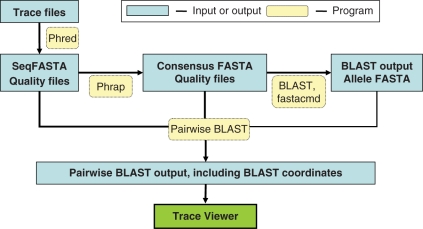
Figure 1.
MLST+ workflow on MGIP. Users upload trace files to the MGIP server for analysis. First, Phred makes base calls on each trace to produce a sequence FASTA file and a quality file. Next, Phrap aligns and produces a consensus sequence FASTA file and other associated files. BLAST is then used to match the consensus sequence against a database of known MLST+ alleles. Allelic FASTA files are extracted from the database using fastacmd and individually aligned to the consensus sequences to determine coordinates, mismatches and indels using pairwise BLAST. Alignments between consensus sequences, called allelic sequences and underlying trace files are displayed using the trace file viewer. The trace file viewer can be used to manually edit consensus sequences based on the aligned trace files (see Figure 3).