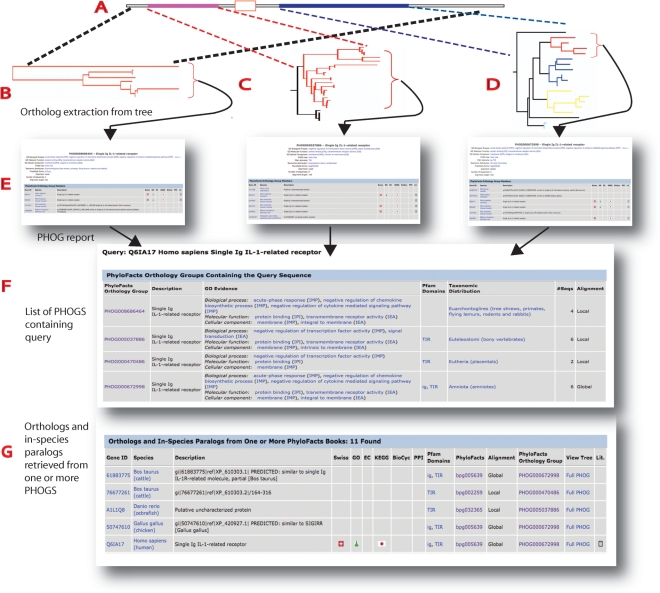
Figure 2.
PhyloFacts ortholog identification pipeline. The input is a protein sequence, in either FASTA format (for BLAST search) or by accession. Results of a sequence accession search are displayed in an Orthology Report including a table of all PHOGs containing the query (F) followed by a table displaying the sequences contained in these PHOGs (G). Links in the columns labelled PhyloFacts Orthology Group retrieve the corresponding PHOG report (E). BLAST results are displayed in an initial table of results (not shown); users would then select one of the sequences in the table, to retrieve the Orthology Report for their selected sequence. (A) Protein sequence query. In this example, the query sequence consists of two evolutionarily conserved domains—an N-terminal Ig domain (pink) followed by a transmembrane helix and and a C-terminal Toll Interleukin Receptor (TIR) domain (blue). (B–D) PhyloFacts trees containing the query sequence are identified, and orthologs are extracted from the orthology group for the sequence (indicated by red subtrees). In this example, the sequence is contained in three PhyloFacts trees. The tree shown in B corresponds to sequences sharing the same overall domain architecture (global homologs). The trees shown in C and D contain sequences that share local (partial) homology along a single domain; the tree in C contains sequences having an Ig domain and the tree in D contains sequences having a TIR domain. (Note that the taxonomic distributions of these PHOGs differ, corresponding to differences in orthology predictions across these domains.) (E) PHOG report—this report displays summary data for the PHOG, followed by a table listing all the orthologs in the PHOG including a link to the sequence database from which the member was drawn, the species of origin, description and links to external resources (e.g. SwissProt, KEGG and BioCyc). (F) List of PHOGs containing the query. This table contains summary data about each PHOG, including PFAM domains, GO annotations and evidence codes and taxonomic distribution. (G) Orthology report: all members of all PhyloFacts orthology groups containing the query are gathered and presented in a table. Note that some orthologs to the query will belong to more than one PHOG (i.e. containing both the ortholog and the query); the column ‘PhyloFacts Orthology Group’ provides a link to the most informative PHOG for each sequence as well as to the PhyloFacts book containing that PHOG. GO annotations and evidence codes, PFAM domains and links to external resources (e.g. SwissProt, KEGG, BioCyc and GO) are also provided. These data are also overlaid on the phylogenetic tree for the PHOG as well as for the family tree from which the PHOG was drawn, and can be viewed using the PhyloScope tree viewer.