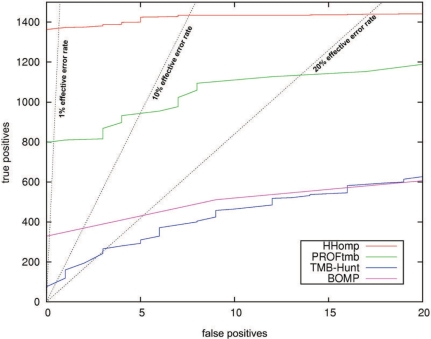
Figure 1.
ROC plot comparing HHomp with the β-barrel prediction methods PROFtmb, TMB-Hunt and BOMP. The true positives (TPs) set consists of the 2164 proteins annotated as OMPs in TransportDB without a BLAST E-value < 0.01 to any of the 23 proteins in our trainings set. The FPs are 5000 randomly selected non-OMP proteins from SCOP. HHomp detects 63.5% of all TPs before the first FP. The effective error rate is the number of effective FPs divided by the sum of TPs and effective FPs are defined as FPs multiplied with 21 to obtain the same fraction of OMPs in the benchmark set as in Gram-negative genomes (2%) (1).