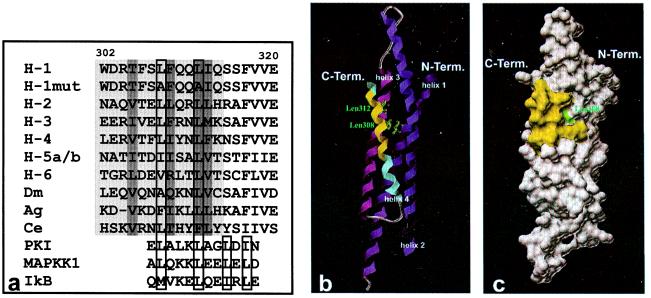
Figure 4.
(a) Aligned sequences of wild-type and NES mutant Stat1 (amino acids 302–320), of homologous regions of Stat family members, and of known leucine-rich export signals of other proteins. The NES sequence of Stat1 between amino acids 302 and 314 and the putative NESs of other Stat proteins are highlighted in light gray. Residues in the interior of the coiled-coil domain are marked with dark gray. Highly conserved hydrophobic residues in position 308 and 312 of the Stat1 NES and important hydrophobic residues in known NES sequences including hallmark residues indicative of the core tetramer are boxed. The following sequences from the National Center for Biotechnology Information Data Library are depicted: H-1, human Stat1, accession no. P42224; H-1mut, human Stat1 mutated in positions 308 and 312; H-2, human Stat2, no. P52630; H-3, human Stat3, no. NP_003141; H-4, human Stat4, no. Q14765; H-5a/b, human Stat5a/b, no. NP_003143/P51692; H-6, human Stat6, no. NP_003144; Dm, Drosophila melanogaster Stat, no. Q24151; Ag, Anopheles gambiae Stat, no. AJ010299; Ce, Caenorhabditis elegans Stat, no. AAD45535; PKI, human cAMP-dependent protein kinase inhibitor α, no. NP_006814; MAPKK1, human mitogen-activated protein kinase kinase 1, no. NP_002746; IkB, human IκBα, no. CAB65556. b and c depict the localization of the NES in the coiled-coil domain (amino acids 136–317) of Stat1. The ribbons drawing (b) shows the four-helix bundle, with amino acids 302–314 of the NES sequence in helix 4 (blue) marked in yellow. The side chains of the leucine residues in position 308 and 312 are included as atomic stick figures. A surface rendering of b is presented in c. The contributions to the surface of the coiled-coil domain by side chains of the NES sequence are highlighted in yellow, with the water accessible area of leucine 308 shown in green.