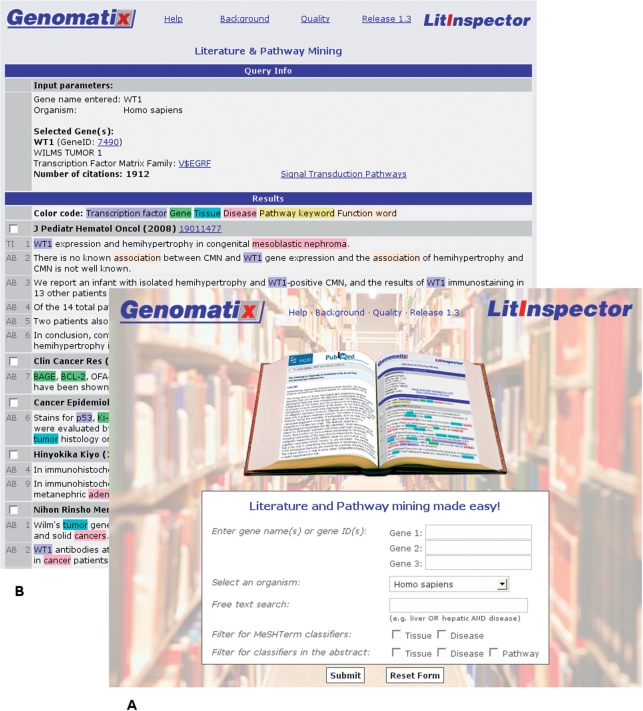
Figure 1.
Screenshot of the LitInspector user interface (A) and a results page (B). (A) The user interface allows input of up to three gene synonyms (e.g. WT1, PAX2) or gene identifiers (e.g. 7490) and free text (e.g. binding site). Default organism is H. sapiens, other organisms can be chosen from a pop-up menu. The search can be filtered for the occurrence of tissue, disease and pathway keywords in the abstract or tissue and disease annotations provided with the MeSH terms. (B) Each result page starts with query info where the user input, the number of citations found and, in case of a gene search, a link to the signal transduction pathway associations is displayed. The results are color-coded sentences wherein genes (green), transcription factors (purple), or keywords (tissue, cyan; disease, pink; pathway, yellow) are highlighted. Genes and transcription factors are hyperlinked to NCBI's Entrez Gene for further information. The output is ordered by the PubMed identifiers (latest publications coming first) which directly link to NCBI's PubMed. The user can select individual abstracts and retrieve them in batch.