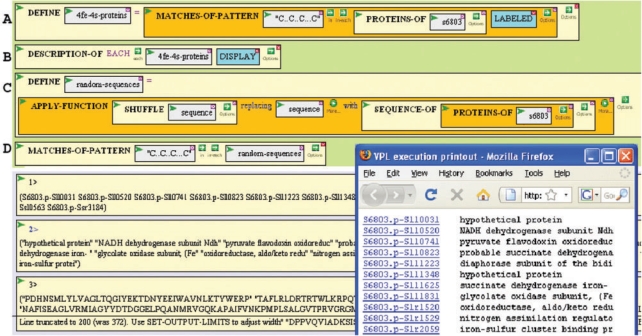
Figure 3.
Example of progressive evaluation and iteration in BioBIKE. The pattern of four cysteine residues separated by 2, 2 and 3 amino acids is often found in proteins with 4Fe-4S clusters (20). (A) The first function finds the pattern of cysteines amongst the sequences of all proteins in the cyanobacterium Synechocystis PCC 6803 and assigns the names of the proteins bearing the motif (Result 1) to a user-defined variable called 4fe-4s-proteins. (B) The annotation for each of the proteins is displayed in a separate window (see inset), and the annotations are also returned as result #2. (C) The user is concerned that this motif might well arise by chance on some proteins of Synechocystis. To test this, a set of random protein sequences is generated, each element being a random shuffling of a real protein sequence. The set is assigned to the variable random-sequences, and the random sequences are returned as result #3. (D) This set of random sequences is searched for the characteristic motif, and none are found (no result), lending some confidence to the belief that the presence of the motif in proteins of Synechocystis is of biological significance.