Abstract
As an increasing number of reliable protein–protein interactions (PPIs) become available and high-throughput experimental methods provide systematic identification of PPIs, there is a growing need for fast and accurate methods for discovering homologous PPIs of a newly determined PPI. PPISearch is a web server that rapidly identifies homologous PPIs (called PPI family) and infers transferability of interacting domains and functions of a query protein pair. This server first identifies two homologous families of the query, respectively, by using BLASTP to scan an annotated PPIs database (290 137 PPIs in 576 species), which is a collection of five public databases. We determined homologous PPIs from protein pairs of homologous families when these protein pairs were in the annotated database and have significant joint sequence similarity (E ≤ 10−40) with the query. Using these homologous PPIs across multiple species, this sever infers the conserved domain–domain pairs (Pfam and InterPro domains) and function pairs (Gene Ontology annotations). Our results demonstrate that the transferability of conserved domain-domain pairs between homologous PPIs and query pairs is 88% using 103 762 PPI queries, and the transferability of conserved function pairs is 69% based on 106 997 PPI queries. The PPISearch server should be useful for searching homologous PPIs and PPI families across multiple species. The PPISearch server is available through the website at http://gemdock.life.nctu.edu.tw/ppisearch/.
INTRODUCTION
Interactions between proteins are critical to most biological processes. To identify and characterize protein–protein interactions (PPIs) and their networks, many high-throughput experimental approaches, such as yeast two-hybrid screening, mass spectroscopy and tandem affinity purification and computational methods [phylogenetic profiles (1), known 3D complexes (2) and interologs (3)] have been proposed (4). Some PPI databases, such as IntAct (5), BioGRID (6), DIP (7), MIPS (8) and MINT (9), have accumulated PPIs submitted by biologists, and those from mining literature, high-throughput experiments and other data sources. As these interaction databases continue growing in size, they become increasingly useful for analysis of newly identified interactions.
The discovery of sequence homologs to a known protein often provides clues for understanding the function of a newly sequenced gene. As an increasing number of reliable PPIs become available, identifying homologous PPIs should be useful to understand a newly determined PPI. Recently, several PPI databases (e.g. IntAct and BioGRID) allow users to input one or a pair of proteins or gene names to acquire the PPIs associated with the query protein(s). Few computational methods (10,11) applied homologous interactions to assess the reliability of PPIs.
To address this issue, we proposed the PPISearch server for searching homologous PPIs across multiple species and annotating the query protein pair. According to our knowledge, PPISearch is the first public server that identifies homologous PPIs from annotated PPI databases and infers transferability of interacting domains and functions between homologous PPIs and the query. PPISearch is an easy-to-use web server that allows users to input a pair of protein sequences. Then, this server finds homologous PPIs in multiple species from five public databases (IntAct, MIPS, DIP, MINT and BioGRID) and annotates the query. Our results demonstrate that this server achieves high agreements on interacting domain–domain pairs and function pairs between query protein pairs and their respective homologous PPIs.
METHOD AND IMPLEMENTATION
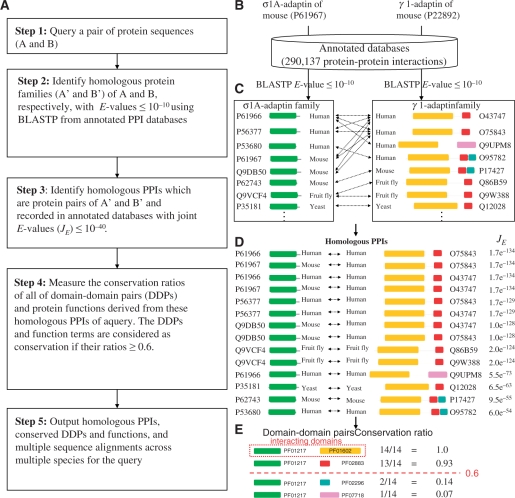
Figure 1 shows the details of the PPISearch server to search homologous PPIs of a query protein pair (A and B) by the following steps (Figure 1A). This server first identifies the homologous families (A′ and B′) of A and B, respectively, with E ≤ 10−10 by using BLASTP to scan the annotated PPI databases (Figure 1B and C). All protein pairs of A′ and B′ are considered candidates of homologous PPIs. We selected homologous PPIs from these candidates, which are recorded in the annotated databases, and have significant joint sequence similarity (E ≤ 10−40) between candidates and the query (Figure 1D). Then, we measure the conservation ratios of domain-domain pairs [DDPs; Pfam (12) and InterPro (13) domains] and protein functions [Gene Ontology annotations (14)] derived from these homologous PPIs of the query (Figure 1E). This server provides conserved DDPs and protein functions for annotating the query. Finally, this server provides homologous PPIs in multiple species; conservations and GO annotations of protein functions; conservations and annotations of DDPs; and the best-matched protein pair of the query.
Figure 1.
Overview of the PPISearch server for homologous protein–protein interaction search and conservation analysis using proteins σ1A-adaptin and γ1-adaptin as the query. (A) The main procedure. (B) Identify homologs of σ1A-adaptin and γ1-adaptin using BLASTP to scan the annotated PPI databases. (C) The homologous families of σ1A-adaptin and γ1-adaptin with E-values ≤10−10. (D) Homologous PPIs of the query. (E) Conservation ratios of domain-domain pairs derived from homologous PPIs.
Homologous protein–protein interaction
The concept of homologous PPI is the core of the PPISearch server to identify the PPI family and measure DDPs and functional conservations of a query protein pair (A and B). We define a homologous PPI as follows: (1) homologs of A and B are proteins with significant sequence similarity BLASTP E-values ≤10−10 (3,15); (2) significant joint sequence similarity (joint E-value JE ≤ 10−40) between two pairs, i.e. (A, A1′) and (B, B1′), of the query protein pair (A and B) and their respective homologs (A1′ and B1′) recorded in annotated PPI databases. This work followed previous studies (3,15) to define joint sequence similarity as
| 1 |
where EA is the E-value of proteins A and A1′; and EB is the E-value of proteins B and B1′. Here, JE ≤ 10−40 is considered a significant similarity according to statistical analysis of 290 137 annotated PPIs and 6597 orthologous PPI families collected from the PORC database (16).
Annotations of homologous PPI
A query protein pair and its homologous PPIs, significant both in sequence and joint sequence similarity, can be considered a PPI family. The concept of PPI families is similar to that of protein sequence family (12,13) and protein structure family (17). We believe that PPI families can be applied widely in biological investigations. Here, we assume that the members of a PPI family are conserved on specific functions and in interacting domain(s). Using these conservations of a PPI family, our server can be used to annotate the protein functions and DDPs of a query protein pair.
Transferability of domain–domain pairs
A query protein pair and its homologous PPIs often show conserve interacting DDPs. To measure the occurence of each DDP in a PPI family, we define the conservation ratio (CRDp) of a DDPp in homologous PPIs of a query protein pair i as
| 2 |
Figure 1D and E show an example to calculate the CRD values of four DDPs. In addition, to evaluate the transferability of DDPs between a query and its homologous PPIs statistically, this study defines the shared ratio (SRD) of DDPs using CRDp and 103 762 annotated PPIs as query protein pairs. The SRD of DDPs against different ratio c is given as
| 3 |
where Q is a set of annotated PPIs in databases (here, the total number of PPIs in Q is 103 762); i is a query protein pair; di(CRDp ≥ c) is the number of DDPs with CRDp values exceeding c; and these DDPs are shared by the query i and its homologous PPIs. Di(CRDp ≥ c) is the total number of the DDPs with CRDp ≥ c, where DDPs are derived from homologous PPIs of the query i. Here, this work used a statistical approach to determine the threshold c (here, c = 0.6) of CRDp to yield reliable DDP annotations with an acceptable level of Di. Please note that CRDp and SRD are computed from a query protein pair and a set of queries, respectively.
Transferability of molecular function
The members of a PPI family often have similar molecular functions. PPISearch uses the molecular function (MF) terms of Gene Ontology (14) to annotate the functions of a query protein pair. The conservation ratio (CRFm) of an MF term pair (MFP) m in homologous PPIs of a query i is utilized to measure the agreement and is defined as
| 4 |
Additionally, the shared ratio of MFPs (SRF), which is statistically derived from 106 997 annotated queries, is utilized to estimate the transferability of conserved function pairs shared by the query and its homologous PPIs. The SRF against different ratio k is defined as
| 5 |
where Q is a set of annotated PPIs in databases; i is a query protein pair; fi(CRFm ≥ k) is the number of MFPs with CRFm values exceeding k and these MFPs are shared by the query i and its homologous PPIs; and Fi(CRFm ≥ k) is the total number of MFPs with CRFm ≥ k, where MFPs are derived from homologous PPIs of the query i. Here, k is set to 0.6.
INPUT, OUTPUT and OPTIONS
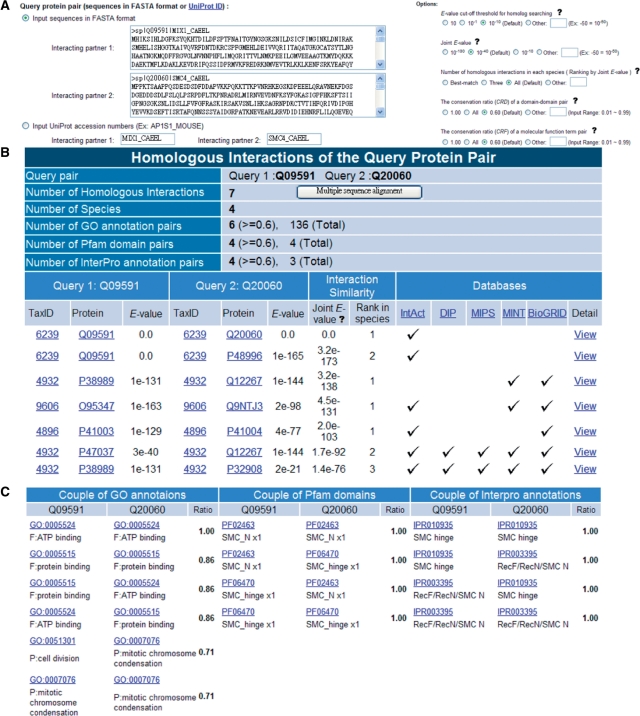
The PPISearch is an easy-to-use web server (Figure 2). Users input a pair of protein sequences in FASTA format or UniProt ID, and choose E-value thresholds for homologs and for homologous PPIs (Figure 2A). In addition, users can assign the CRD and CRF thresholds, specific species and the number of homologous PPIs in a species.
Figure 2.
The PPISearch server search results using proteins MIX-1 and SMC-4 of Caenorhabditis elegans as the query. (A) The user interface for assignments of query protein sequences and E-value thresholds of homologs and homologous PPIs. (B) Homologous PPIs of MIX-1−SMC-4 in multiple species and public databases. (C) Conserved protein functions (GO terms) and domain-domain pairs (Pfam and InterPro) of homologous PPIs with a conservation ratio ≥ 0.6.
Typically, the PPISearch server yields homologous PPIs within 20 s when sequence length is ≤350 (Figure 2B). This server identifies homologous PPIs in multiple species; conservations and GO annotations of protein functions; conservations and annotations of DDPs; and the best-matched protein pairs of the query (Figure 2C). Additionally, the PPISearch server provides multiple sequence alignments of homologous PPIs and indicates the conserved residues based on amino acid types. For each homologous PPI, this server shows the alignments and experimental annotations (e.g. interaction types, experimental methods, gene names and GO terms).
Example analysis
σ1A-adaptin and γ1-adaptin
Figure 1C and D show search results using σ1A-adaptin (UniProt accession number: P61967) and γ1-adaptin (P22892) of Mus musculus as the query. These two proteins are components of the heterotetrameric adaptor protein complex 1 (AP-1), which medicates clathrin-coated vesicle transport from the trans-Golgi network to endosome (18). According to the crystal structure (PDB code 1W63) (19), this protein pair is a physical interaction, but it is not recorded in the annotated PPI database. For this query, the PPISearch server identifies 14 homologous PPIs, a PPI family, from four species (human, mouse, fruit fly and yeast). This PPI family has four DDPs (Figure 1E)—PF01217-PF01602 (CRD is 1.0), PF01217-PF02883 (0.93), PF1217-PF02296 (0.14) and PF01217-PF07718 (0.07). Two DDPs (PF01217-PF01602 and PF01217-PF02883) with highest CRD ratios are the domain compositions of the query and PF01217-PF01602 is the interacting domains (19).
This server allows users to choose the JE threshold of homologous PPIs. For example, when JE is set to 10−100 (default value is 10−40), the number of homologous PPIs decreases from 14 to 10 by filtering out the last four PPIs (Figure 1D). These 10 homologous PPIs consistently include the two DDPs PF01217-PF01602 and PF01217-PF02883, each with a CRD = 1.0. Furthermore, users can choose the best match or number of homologous PPIs in a species. In this manner, the PPISearch server is able to select the primary homologous PPIs of each species for specific applications, such as evolutionary analysis of essential proteins.
MIX-1 and SMC-4
Mitotic chromosome and X-chromosome-associated protein (MIX-1, Q09591) and structural maintenance of chromosomes protein 4 (SMC-4, Q20060) of Caenorhabditis elegans are members of SMC protein family, and are required for mitotic chromosome segregation (20). Both MIX-1 and SMC-4 are essential components in forming the condensin complex for interphase chromatin to convert into mitotic-like condense chromosomes (20,21). Using C. elegans MIX-1 and SMC-4 as the query protein pair and JE is set to 10−40, the PPISearch server found seven homologous interactions from annotated PPI databases (Figure 2B). These seven homologous PPIs are consistently SMC–SMC protein interactions, including SMC-2−SMC-4, SMC-3−SMC-4 and SMC-2−SMC-1, in four species. Among these homologous PPIs, two PPIs, Q95347-Q9NTJ3 (Homo sapiens) and P38989-Q12267 (Saccharomyces cerevisiae), are orthologous interactions of the query MIX-1−SMC-4 (16).
These seven homologous PPIs of MIX-1 and SMC-4 include 136 GO term pairs. Among these GO terms, the CRF ratios of four GO MF term pairs and two GO BP term pairs exceed 0.6 (Figure 2C). These six GO term pairs are consistent with the term-pair combinations of MIX-1 and SMC-4. For example, MIX-1 and SMC-4 have the same two GO MF annotations, protein binding (GO:0005515) and ATP-binding (GO:0005524). Additionally, these seven homologous PPIs contain four DDPs with CRD ratios of 1.0. These four DDPs, PF02463-PF02463, PF06470-PF02463, PF02463-PF06470 and PF06470-PF06470, are recorded in iPfam (12) and are consistent with the query pair. The hinge–hinge interaction (PF02463-PF02463) is experimentally proved, and is conserved in the eukaryotic SMC-2–SMC-4 heterodimer (22). These analytical results reveal that the PPISearch server is able to identify homologous PPIs that share conserved DDPs and MFPs with the query.
RESULTS
To evaluate the usefulness of the PPISearch server for the discovery of homologous PPIs and for the annotations of a query protein pair, we selected two query protein sets, termed HOM and ORT. To search homologous PPIs, HOM and ORT are used to assess PPISearch performance and to determine the threshold of joint E-value JE [Equation (1)] (Figure 3A). In addition, the HOM set was applied to infer the relations between conservation ratios [CRD and CRF defined in Equations (2) and (4)] and the transferability of DDPs and MFPs, respectively, between a query and its homologous PPIs (Figure 3B and Supplementary Figure S1). The HOM set includes all 290 137 PPIs and the ORT set has 6597 orthologous PPI families (14 571 PPIs) derived from the annotated PPI database and PORC orthology database (16).
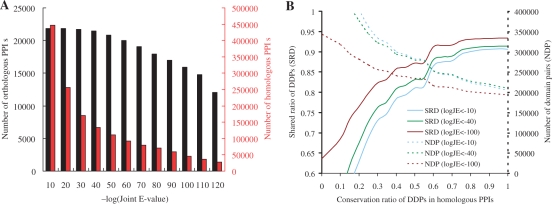
Figure 3.
Evaluations of the PPISearch server. (A) The relationships between joint E-value JE and the numbers of orthologous PPIs (black) and homologous PPIs (red) derived from 290 137 annotated PPIs. (B) The relationships between conservation ratios of DDPs with shared ratios of DDPs and with the number (dotted lines) of DDPs derived from 103 762 PPI families. The shared ratio of DDPs is 0.88 and the number of DDPs is 252 728 when the conservation ratio is 0.6 and joint E-value is 10−40 (green lines).
HOM and ORT were used to assess the PPISearch server in identifying homologous PPIs and orthologous PPIs, respectively, by searching the annotated PPI database (290 137 PPIs with 54 422 proteins). Figure 3A shows the relationships between joint E-value JE and number of orthologous PPIs (black) and homologous PPIs (red). The orthologous PPIs often have the same functions and domains. When JE ≤ 10−40, the number of orthologous PPIs decreases significantly; conversely, the number of homologous PPIs decreases more gradually than that at JE ≥ 10−40. This result shows that the proposed method is able to identify 98.2% orthologous PPIs with a reasonable number of homologous PPIs when JE ≤ 10−40.
To evaluate the transferability of DDPs and MFPs between a query and its homologous PPIs, we used the SRD [Equation (3)] and SRF [Equation (5)]. The HOM set is used to evaluate the utility of the PPISearch server in annotating the query protein pair. By excluding proteins without domain annotations from the query set, 103 762 PPIs are used to evaluate the transferability (SRD) of conserved DDPs between these query PPIs and their respective homologous PPIs (Figure 3B). The transferability (SRF) of conserved functions between the 106 997 PPIs and their homologous PPIs is assessed by excluding proteins without molecular function terms of GO from the original query set (Supplementary Figure S1).
Figure 3B shows the relationship between conservation ratios (CRD) of DDPs and the SRD ratios. The SRD ratio increases significantly (solid lines) when the CRD increases and CRD ≤ 0.6. Conversely, the number of DDPs derived from 103 762 PPI families decreases (dotted lines) as CRD increases. If the CRD is set to 0.6 and the joint E-value is set to 10−40 (green lines), the SRD is 0.88 and the number of DDPs is 252 728. This result demonstrates that members of a PPI family derived by PPISearch reliably share DDPs (or interacting domains). Additionally, similar results were obtained for transferability of conserved functions between homologous PPIs and the query (Supplementary Figure S1). The members of a PPI family have similar molecular functions, and SRF ratios are highly correlated with conservation ratios (CRF) of MFPs. When the CRF is 0.6 and the joint E-value is 10−40 (green lines), the SRF is 0.69 and the number of MFPs is 454 251.
These results reveal that the PPISearch server achieves a high SRD with a reasonable number of DDPs when the joint E-value is set to 10−40. In summary, these experimental results demonstrate that this server achieves high agreement on DDPs and MFPs between the query and their respective homologous PPIs.
CONCLUSIONS
This study demonstrates the utility and feasibility of the PPISearch server in identifying homologous PPIs and inferring conserved DDPs and MFPs from PPI families. By allowing users to input a pair of protein sequences, PPISearch is the first server that can identify homologous PPIs from annotated PPI databases and infer transferability of interacting domains and functions between homologous PPIs and a query. Our experimental results demonstrate that the query protein pair and its homologous PPIs achieve high agreement on conserved DDPs and MFPs. We believe that PPISearch is a fast homologous PPIs search server and is able to provide valuable annotations for a newly determined PPI.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council and partial support of the ATU plan by MOE to J.-M.Y. Funding for open access charge: National Science Council of the Republic of China and MOE ATU.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Authors are grateful to both the hardware and software supports of the Structural Bioinformatics Core Facility at National Chiao Tung University.
REFERENCES
- 1.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y-C, Lo Y-S, Hsu W-C, Yang J-M. 3D-partner: a web server to infer interacting partners and binding models. Nucleic Acids Res. 2007:W561–W567. doi: 10.1093/nar/gkm346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu HY, Luscombe NM, Lu HX, Zhu XW, Xia Y, Han JDJ, Bertin N, Chung S, Vidal M, Gerstein M. Annotation transfer between genomes: Protein-protein interologs and protein-DNA regulogs. Gen. Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLoS Comput. Biol. 2007;3:337–344. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct – open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mewes HW, Dietmann S, Frishman D, Gregory R, Mannhaupt G, Mayer KFX, Munsterkotter M, Ruepp A, Spannagl M, Stuempflen V, et al. MIPS: analysis and annotation of genome information in 2007. Nucleic Acids Res. 2008;36:D196–D201. doi: 10.1093/nar/gkm980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatr-Aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patil A, Nakamura H. Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC Bioinformatics. 2005;6:100–112. doi: 10.1186/1471-2105-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saeed R, Deane C. An assessment of the uses of homologous interactions. Bioinformatics. 2008;24:689–695. doi: 10.1093/bioinformatics/btm576. [DOI] [PubMed] [Google Scholar]
- 12.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Gen. Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, Phan I, et al. Integr8 and genome reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Ann. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 19.Heldwein EE, Macia E, Jing W, Yin HL, Kirchhausen T, Harrison SC. Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl Acad. Sci. USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: an essential component of the C. elegans mitotic machinery executes x chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 21.Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.