Abstract
The RosettaAntibody server (http://antibody.graylab.jhu.edu) predicts the structure of an antibody variable region given the amino-acid sequences of the respective light and heavy chains. In an initial stage, the server identifies and displays the most sequence homologous template structures for the light and heavy framework regions and each of the complementarity determining region (CDR) loops. Subsequently, the most homologous templates are assembled into a side-chain optimized crude model, and the server returns a picture and coordinate file. For users requesting a high-resolution model, the server executes the full RosettaAntibody protocol which additionally models the hyper-variable CDR H3 loop. The high-resolution protocol also relieves steric clashes by optimizing the CDR backbone torsion angles and by simultaneously perturbing the relative orientation of the light and heavy chains. RosettaAntibody generates 2000 independent structures, and the server returns pictures, coordinate files, and detailed scoring information for the 10 top-scoring models. The 10 models enable users to use rational judgment in choosing the best model or to use the set as an ensemble for further studies such as docking. The high-resolution models generated by RosettaAntibody have been used for the successful prediction of antibody–antigen complex structures.
INTRODUCTION
Therapeutic monoclonal antibodies are a genre of biopharmaceuticals which has benefitted healthcare in various fields from oncology to immune and inflammatory disorders. Development of successful novel therapeutic antibodies requires understanding of drug and disease mechanisms and the ability to stabilize, affinity mature, and humanize antibodies. Antibody structures can help overcome these challenges by providing atomic level insights into structure–function relationships and the antibody–antigen interaction [e.g. see refs. (1–4)]. However, experimental techniques for obtaining antibody structures, like X-ray crystallography and nuclear magnetic resonance, are laborious, time consuming and costly. Computational antibody structure prediction provides a fast and inexpensive route to obtain structures, including those which are not obtainable otherwise.
Two antibody variable region (FV) modeling servers are available on the Internet: the Web Antibody Modeling (WAM) (5) and Prediction of Immunoglobulin Structure (PIGS) (6) servers. WAM can require several days to output one antibody model in response to a submitted query sequence. No information on templates used for modeling the antibody is provided. Furthermore, antibody structures predicted with WAM have internal clashes and their inaccuracies can confound computational docking (2,7). The PIGS server returns an antibody model in about a minute and displays the antibody crystal structures that it selects as templates. The PIGS models are generated by grafting complementarity determining region (CDR) loops onto selected framework templates, even for the hyper-variable and non-canonical CDR H3 loop. Accurate CDR H3 predictions would only be expected when a similar CDR H3 loop is present in the database, which is unlikely for novel antibody sequences. The existing servers do not provide high-resolution refinement of antibody structures and do not consider thermodynamics during modeling.
RosettaAntibody (7) is a homology modeling program within the Rosetta suite (8) for predicting high-resolution antibody FV structures. The prediction includes modeling CDR H3 loop conformations, and it uses a simple free energy function to relieve steric clashes by simultaneously optimizing the CDR loop backbone dihedral angles, the relative orientation of the light (VL) and heavy (VH) chains, and the side chain conformations. A crude model where all the CDRs are grafted from template structures can be provided in a few minutes, and high-resolution models can be generated by the full RosettaAntibody protocol running on a cluster of computers in about a day (sometimes users may need to wait for other jobs in the queue). The 10 top-scoring RosettaAntibody models can be used in docking techniques such as EnsembleDock (9) that can select binding competent conformers during docking.
A few limitations of RosettaAntibody have been that (i) execution of multiple scripts for the identification of templates can be complex, (ii) finding the template structures that have been used in modeling can be challenging given the large number of intermediate files that are generated (iii) the Rosetta command-line interface can be difficult to use and (iv) it requires significant computational time to generate all-atom models, requiring a cluster of computers. To overcome these limitations and to make the high-resolution modeling available to a broader community, we have developed the RosettaAntibody server (http://antibody.graylab.jhu.edu), where the interface is simple and modest computing resources are provided.
PROCESSING METHOD
RosettaAntibody predicts the structure of the FV region in two stages. The first stage identifies the CDR loops and the framework regions in the input sequences, chooses the most sequence homologous templates for each respective segment, grafts the template CDRs onto template frameworks, and finally optimizes the side chains of all the residues in the assembled model. The crude model generated in the first stage is used as an input to the second stage.
The second stage of RosettaAntibody is a multi-start, multi-scale Monte Carlo-plus-minimization algorithm that generates two thousand candidate structures. The second stage of the algorithm is split into a low-resolution and a high-resolution phase. The low-resolution phase represents side chains as single pseudo-atoms (10) and generates candidate CDR H3 loop conformations via fragment assembly and cyclic coordinate descent (11) in a Monte Carlo loop. Scoring in the low-resolution phase favors nonlocal properties of native protein structures such as hydrophobic burial, compactness, pairing of β-strands and closure of the chain gap during loop building (12). The high-resolution phase iteratively performs the following: (i) optimizes side chains via rotamer packing and continuous minimization (13), (ii) perturbs CDR backbone torsion angles and the relative orientation of the light and heavy chains, and (iii) uses gradient-based minimization over the CDR torsion angles and the light chain-heavy chain displacement. The high-resolution energy function includes van der Waals energy, orientation-dependent hydrogen bonding (14), implicit Gaussian solvation (15), side-chain rotamer propensities (16), and a low-weighted distance-dependent dielectric electrostatic energy (17). Complete methodological details are provided in ref. (7).
INPUTS AND OUTPUTS
Input
Amino-acid sequences of the light and heavy chains of the FV region are submitted to the server by either pasting the sequences in the appropriate field or by uploading them as two separate FASTA formatted files. Since the RosettaAntibody server models only the FV region of the antibody, FC and leader sequences should be truncated from the input sequence prior to submission. To uniquely identify each job, the user must specify the name of the antibody and can optionally specify the user's name and an email address for notification when the modeling task has finished.
Output
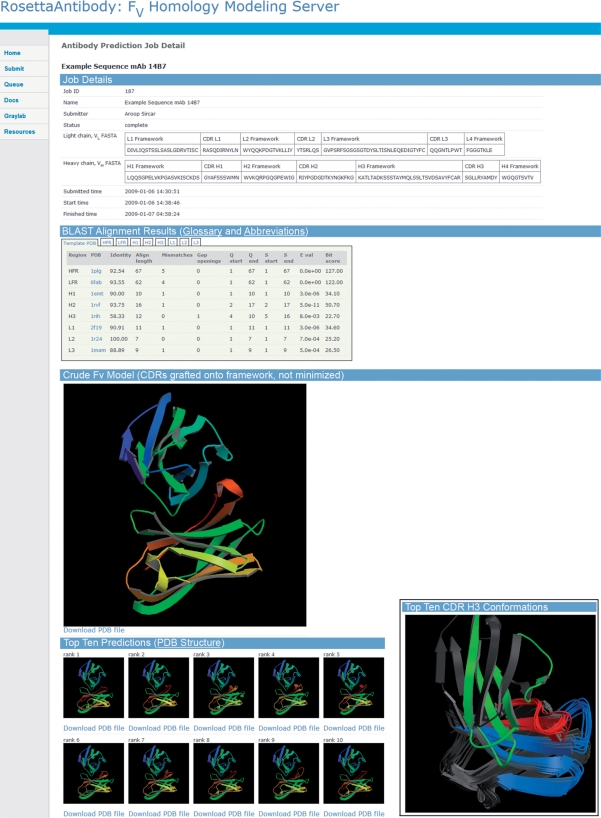
Figure 1 shows a representative output page from the RosettaAntibody server. The top of the page summarizes the details of the respective job, e.g. the name of the antibody, the name of the user (if provided), and the date and time of submission, execution and completion. A chart shows the boundaries of the CDR loops and the framework regions. Next, a table displays the most sequence-homologous template [identified by BLAST (18)] for each antibody segment (VH and VL frameworks, CDRs L1, L2, L3, H1, H2 and H3). In a series of selectable panes, the table also displays the top seven templates for each antibody segment. A picture of the crude FV model, formed by joining the top templates, is shown next on the output page with a link for downloading a file containing the coordinates and various energies. Residue numbering of all models generated by RosettaAntibody follows Chothia's antibody numbering scheme (19).
Figure 1.
Sample results page provided by the server for monoclonal antibody 14B7 (49). Inset shows the overlay of the 10 top-scoring RosettaAntibody models which appears further down the page (antibody framework regions, gray; light chain CDRs, green; CDRs H1 and H2, blue; CDR H3, red). Web page images are generated with MolScript (50).
If requested, the next section of the page will show the 10 top-scoring high-resolution structures in rank order by energy. Each model output file includes the scoring data of individual energy terms (van der Waals, solvation, hydrogen bonding energies, etc.) for the whole FV model as well as residue-by-residue breakdowns. Finally, the web page shows a picture of a superposition of the 10 top-scoring structures from the perspective of an antigen, demonstrating the differences in the different models (figure inset).
Any difficulties in processing the sequence would be shown in a ‘warnings’ section. Warnings sometimes arise to indicate poor matching of template structures or broken predicted CDR H3 loop conformations, resulting in a lower confidence model. Additional explanations of the warnings are provided in the website documentation. The documentation page also explains all output in detail, including a description of the scoring terms found in the coordinate files.
SYSTEM ARCHITECTURE
The RosettaAntibody server has a front-end web process which interfaces with the computation daemon and engine. The front-end, implemented in Python using Django (http://www.djangoproject.com), provides results upon request for users and enters modeling tasks into a MySQL database once an input file is submitted. A back-end daemon pulls tasks from the queue in the MySQL database, translates the modeling task into Perl wrapper scripts that detect the different segments of the antibody variable region, runs BLAST to detect templates, specifies a Rosetta++ command-line, and finally submits a job to a Condor (http://www.cs.wisc.edu/condor) queue. Condor runs the job on our 188-processor Linux cluster at Johns Hopkins University, as time is available. The back-end daemon periodically detects the status of the job to report, and eventually enters the complete set of results into the MySQL database.
SERVER PERFORMANCE
Since the RosettaAntibody web server opened in December of 2007, over 100 individuals have used the web server for more than 400 modeling jobs. Jobs typically require about 1700 processor-hours, and results are typically complete within a day of submission, although the time will vary with the current cluster load and server queue. The website is free and open to all users with no login requirement.
Validation of the RosettaAntibody server algorithm
In a large scale test of RosettaAntibody, the program was used to recover the native crystal structures of 54 antibodies (7). To simulate blind prediction, when database information was used in modeling, only nonrelated (less than 90% sequence identity) antibody structures in the Protein Data Bank (20) were used. For the best ranked model of each target, the median root mean square deviation (rmsd) of the antigen binding pocket comprising of all the CDR residues was 1.5 Å, and 80% of the targets had an rmsd lower than 2.0 Å.
The loop modeling capabilities of RosettaAntibody were tested by ab initio modeling of the CDR H3 loop. The CDR H3 loop is composed of residues 95–102 of the heavy chain [Chothia numbering (19)]. The median backbone heavy atom global rmsd of the CDR H3 loop prediction for the best ranked model was 1.6, 1.9, 2.4, 3.1 and 6.0 Å, respectively, for very short (4–6 residues), short (7–9 residues), medium (10–11 residues), long (12–14 residues) and very long (17–22 residues) loops.
Finally, a practical measure of the accuracy of the antibody structures is their utility for docking to antigens. While the inclusion of the RosettaAntibody refinement steps had a small effect on homology modeling rmsds (other than CDR H3), refinement was critical for achieving docking accuracy (7). When the set of 10 top-scoring RosettaAntibody FV homology models was used in local ensemble docking to antigen, a moderate-to-high accuracy docking prediction [rated by Critical Assessment of PRediction of Interactions criteria (21)] was achieved in 7 of 15 targets (7).
In a comparison of WAM and RosettaAntibody (7), for some antibodies, the CDR H3 predicted by WAM was closer to the native structure than that of the top-scoring model produced by RosettaAntibody. However, there was typically a more accurate structure among the 10 top-scoring RosettaAntibody models. Furthermore, antibody–antigen docking simulations starting with RosettaAntibody FV models consistently resulted in more accurate docking predictions than those obtained by starting with WAM generated models or unrefined RosettaAntibody models (7).
Potential uses of the RosettaAntibody server
Antibody structures can be used to guide rational efforts to enhance stability (22,23) or to humanize sequences to minimize immunological response (24,25). Antibody structures can also be used for docking to their antigens, either for epitope mapping (26) or for high-resolution refinement (27). For example, we docked models of monoclonal antibody 14B7 to the anthrax toxin protective antigen (2). The models helped us form hypotheses about the mechanism of affinity maturation of several variants of 14B7. Several other instances of docking antibody homology models are present in the literature (28–30). Docking calculations can be done on several publicly available servers (31–38) including the RosettaDock Server (local docking only for high-resolution refinement, http://rosettadock.graylab.jhu.edu) (39).
Docking of homology models is necessarily less accurate than docking of crystal structures. Experimental information can be used to mitigate errors. For example, we used computational mutagenesis and hotspot analysis to evaluate models of epidermal growth factor receptor binding to mAb 806 (1). In recognition of the errors present in homology models, RosettaAntibody provides 10 alternate low-energy structures. There are several new docking methods which can use multiple input structures for one of the docking partners (40–43). Our EnsembleDock program (9) can improve low-energy docking solutions, and sometimes the low-energy docking solution is formed by the component homology structure that is closest to the crystal structure.
FUTURE DIRECTIONS
Accurate loop modeling remains one of the central challenges of antibody modeling. Thus, future improvements might be made as better and more efficient loop modeling algorithms [e.g. kinematic loop closure (44) or hierarchical local optimization (45,46)] become available. Predictions might also be improved by inclusion of NMR constraints to bias simulations [e.g. (47)]. Some researchers are pursuing therapeutics based ‘heavy chain only’ (VHH) antibodies discovered in the blood of camelids (48). VHHs are also easy to clone and express, and their structure might be amendable to prediction, although tests are required to assess the use of standard antibody database. Finally, we are currently developing flexible backbone antibody docking techniques which exploit the same antibody structural modeling tools as the server. These induced-fit antibody docking techniques may additionally help overcome homology modeling errors as predicted structures are used for high-resolution applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01-GM073151, R01-GM078221) and UCB S.A. Funding for open access charge: National Institutes of Health Grant Number R01-GM078221.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Sergey Lyskov for technical help in programming the server and Jan Vandenberg and Alainna White for administration of their network and servers.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Sivasubramanian A, Chao G, Pressler HM, Wittrup KD, Gray JJ. Structural model of the mAb 806-EGFR complex using computational docking followed by computational and experimental mutagenesis. Structure. 2006;14:401–414. doi: 10.1016/j.str.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 2.Sivasubramanian A, Maynard JA, Gray JJ. Modeling the structure of mAb 14B7 bound to the anthrax protective antigen. Proteins. 2008;70:218–230. doi: 10.1002/prot.21595. [DOI] [PubMed] [Google Scholar]
- 3.Lippow SM, Wittrup KD, Tidor B. Computational design of antibody-affinity improvement beyond in vivo maturation. Nat. Biotechnol. 2007;25:1171–1176. doi: 10.1038/nbt1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariuzza RA, Phillips SE, Poljak RJ. The structural basis of antigen-antibody recognition. Annu. Rev. Biophys. Biophys. Chem. 1987;16:139–159. doi: 10.1146/annurev.bb.16.060187.001035. [DOI] [PubMed] [Google Scholar]
- 5.Whitelegg NR, Rees AR. WAM: an improved algorithm for modelling antibodies on the WEB. Protein Eng. 2000;13:819–824. doi: 10.1093/protein/13.12.819. [DOI] [PubMed] [Google Scholar]
- 6.Marcatili P, Rosi A, Tramontano A. PIGS: automatic prediction of antibody structures. Bioinformatics. 2008;24:1953–1954. doi: 10.1093/bioinformatics/btn341. [DOI] [PubMed] [Google Scholar]
- 7.Sivasubramanian A, Sircar A, Chaudhury S, Gray JJ. Toward high-resolution homology modeling of antibody Fv regions and application to antibody-antigen docking. Proteins. 2009;74:497–514. doi: 10.1002/prot.22309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das R, Baker D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhury S, Gray JJ. Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles. J. Mol. Biol. 2008;381:1068–1087. doi: 10.1016/j.jmb.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 11.Canutescu AA, Dunbrack R.L., Jr. Cyclic coordinate descent: a robotics algorithm for protein loop closure. Protein Sci. 2003;12:963–972. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang C, Schueler-Furman O, Baker D. Improved side-chain modeling for protein-protein docking. Protein Sci. 2005;14:1328–1339. doi: 10.1110/ps.041222905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kortemme T, Morozov AV, Baker D. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 2003;326:1239–1259. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 15.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 16.Dunbrack RL, Jr., Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warshel A, Russell ST, Churg AK. Macroscopic models for studies of electrostatic interactions in proteins: limitations and applicability. Proc. Natl Acad. Sci. USA. 1984;81:4785–4789. doi: 10.1073/pnas.81.15.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J. Mol. Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 20.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3-5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 22.Honegger A, Malebranche AD, Rothlisberger D, Pluckthun A. The influence of the framework core residues on the biophysical properties of immunoglobulin heavy chain variable domains. Protein Eng. Des. Sel. 2009;22:121–134. doi: 10.1093/protein/gzn077. [DOI] [PubMed] [Google Scholar]
- 23.Clark LA, Boriack-Sjodin PA, Eldredge J, Fitch C, Friedman B, Hanf KJ, Jarpe M, Liparoto SF, Li Y, Lugovskoy A, et al. Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design. Protein Sci. 2006;15:949–960. doi: 10.1110/ps.052030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlapschy M, Fogarasi M, Gruber H, Gresch O, Schafer C, Aguib Y, Skerra A. Functional humanization of an anti-CD16 Fab fragment: obstacles of switching from murine λ to human λ or κ light chains. Protein Eng. Des. Sel. 2008;22:175–188. doi: 10.1093/protein/gzn066. [DOI] [PubMed] [Google Scholar]
- 25.Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986;321:522–525. doi: 10.1038/321522a0. [DOI] [PubMed] [Google Scholar]
- 26.Ponomarenko JV, Bourne PE. Antibody-protein interactions: benchmark datasets and prediction tools evaluation. BMC Struct. Biol. 2007;7:64. doi: 10.1186/1472-6807-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray JJ. High-resolution protein-protein docking. Curr. Opin. Struct. Biol. 2006;16:183–193. doi: 10.1016/j.sbi.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Autore F, Melchiorre S, Kleinjung J, Morgan WD, Fraternali F. Interaction of malaria parasite-inhibitory antibodies with the merozoite surface protein MSP1(19) by computational docking. Proteins. 2007;66:513–527. doi: 10.1002/prot.21212. [DOI] [PubMed] [Google Scholar]
- 29.Bracci L, Pini A, Bernini A, Lelli B, Ricci C, Scarselli M, Niccolai N, Neri P. Biochemical filtering of a protein-protein docking simulation identifies the structure of a complex between a recombinant antibody fragment and alpha-bungarotoxin. Biochem. J. 2003;371:423–427. doi: 10.1042/BJ20021369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinney BA, Kallewaard NL, Crowe J.E., Jr., Meiler J. Using the natural evolution of a rotavirus-specific human monoclonal antibody to predict the complex topography of a viral antigenic site. Immunome Res. 2007;3:8. doi: 10.1186/1745-7580-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 32.Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34:W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 34.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 36.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–W232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesk VI, Sternberg MJ. 3D-Garden: a system for modelling protein-protein complexes based on conformational refinement of ensembles generated with the marching cubes algorithm. Bioinformatics. 2008;24:1137–1144. doi: 10.1093/bioinformatics/btn093. [DOI] [PubMed] [Google Scholar]
- 38.Terashi G, Takeda-Shitaka M, Takaya D, Komatsu K, Umeyama H. Searching for protein-protein interaction sites and docking by the methods of molecular dynamics, grid scoring, and the pairwise interaction potential of amino acid residues. Proteins. 2005;60:289–295. doi: 10.1002/prot.20572. [DOI] [PubMed] [Google Scholar]
- 39.Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–W238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grunberg R, Leckner J, Nilges M. Complementarity of structure ensembles in protein-protein binding. Structure. 2004;12:2125–2136. doi: 10.1016/j.str.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 41.Smith GR, Sternberg MJ, Bates PA. The relationship between the flexibility of proteins and their conformational states on forming protein-protein complexes with an application to protein-protein docking. J. Mol. Biol. 2005;347:1077–1101. doi: 10.1016/j.jmb.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 42.Bastard K, Prevost C, Zacharias M. Accounting for loop flexibility during protein-protein docking. Proteins. 2006;62:956–969. doi: 10.1002/prot.20770. [DOI] [PubMed] [Google Scholar]
- 43.Krol M, Chaleil RA, Tournier AL, Bates PA. Implicit flexibility in protein docking: cross-docking and local refinement. Proteins. 2007;69:750–757. doi: 10.1002/prot.21698. [DOI] [PubMed] [Google Scholar]
- 44.Coutsias EA, Seok C, Jacobson MP, Dill KA. A kinematic view of loop closure. J. Comput. Chem. 2004;25:510–528. doi: 10.1002/jcc.10416. [DOI] [PubMed] [Google Scholar]
- 45.Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 46.Zhu K, Pincus DL, Zhao S, Friesner RA. Long loop prediction using the protein local optimization program. Proteins. 2006;65:438–452. doi: 10.1002/prot.21040. [DOI] [PubMed] [Google Scholar]
- 47.Meiler J, Baker D. Rapid protein fold determination using unassigned NMR data. Proc. Natl Acad. Sci. USA. 2003;100:15404–15409. doi: 10.1073/pnas.2434121100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roovers RC, van Dongen GA, en Henegouwen PM. Nanobodies in therapeutic applications. Curr. Opin. Mol. Ther. 2007;9:327–335. [PubMed] [Google Scholar]
- 49.Maynard JA, Maassen CB, Leppla SH, Brasky K, Patterson JL, Iverson BL, Georgiou G. Protection against anthrax toxin by recombinant antibody fragments correlates with antigen affinity. Nat. Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- 50.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]