Abstract
Synthetic Biology, a multidisciplinary field, is growing rapidly. Improving the understanding of biological systems through mimicry and producing bio-orthogonal systems with new functions are two complementary pursuits in this field. A web server called FMM (From Metabolite to Metabolite) was developed for this purpose. FMM can reconstruct metabolic pathways form one metabolite to another metabolite among different species, based mainly on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and other integrated biological databases. Novel presentation for connecting different KEGG maps is newly provided. Both local and global graphical views of the metabolic pathways are designed. FMM has many applications in Synthetic Biology and Metabolic Engineering. For example, the reconstruction of metabolic pathways to produce valuable metabolites or secondary metabolites in bacteria or yeast is a promising strategy for drug production. FMM provides a highly effective way to elucidate the genes from which species should be cloned into those microorganisms based on FMM pathway comparative analysis. Consequently, FMM is an effective tool for applications in synthetic biology to produce both drugs and biofuels. This novel and innovative resource is now freely available at http://FMM.mbc.nctu.edu.tw/.
INTRODUCTION
Synthetic Biology, a rapidly growing multidisciplinary field, has two complementary goals- further elucidating biological systems through mimicry and producing bio-orthogonal systems with new functions (1). Microbes are well established as effective hosts for the biosynthesis of bio-molecules; consequently, engineered microbial biological systems represent critical frontier in synthetic biology (2). Engineered microorganisms are employed in numerous applications, including food additives, pharmaceuticals, fuels, animal feed supplements, cosmetics and polymer materials (3,4). For example, Corynebacterium glutamicum and Escherichia coli are used to produce lysine, methionine, valine and threonine (3,5–8) which are essential intermediate precursors for antibiotics (9) and biofuels (10). These benefits have motivated scientists to design and construct new biological components (such as enzymes, genetic circuits, metabolic pathways and cells) and redesign existing biological systems (11). Furthermore, synthetic biology can be modeled, understood, and tuned to meet particular performance criteria, and smaller components can be assembled into larger integrated systems that solve specific problems (11). Hence, more bioinformatics tools have been created recently for applications in metabolic engineering.
Since 1995, Kanehisa et al. has been developing knowledge-based methods for elucidating higher-order systemic behaviors of cells and organisms from genomic and molecular information (12–15). They established the Kyoto Encyclopedia of Genes and Genomes (KEGG) database for integrating genomic, chemical, metabolic pathways, and systemic functional information (12–15). MetaCyc (16) is a non-redundant database and contains more than 1100 experimentally elucidated metabolic pathways from more than 1500 organisms; it provides a set of reference data for computationally predicting metabolic pathways. Several important databases of metabolic pathways exist with hundreds of metabolic pathways and thousands of biochemical reactions; even the metabolic pathways for a small organism constitutes a large network, such as Roche Applied Science Biochemical Pathways chart (17) and BRENDA (18). Although numerous servers can reconstruct metabolic pathways based on available metabolic maps, it is still inconvenient to reconstruct metabolic pathways from two interesting metabolites. For instance, determining the metabolic pathway from glucose to pyruvate from those databases is arduous unless the two metabolites are already known to be involved in the glycolysis pathway. Moreover, only one metabolite can be input to any of those servers, and users need considerable time to find a specific pathway in which they are interested. In addition, comparative analysis of metabolic networks in various species yields important information for many topics in biology and life sciences, such as evolution and speciation, metabolic engineering, and drug design.
Li et al. (19) adopted Metabolic Pathway Alignment and Scoring (M-PAS) for identifying and ranking conserved metabolic pathways based on a comprehensive and flexible similarity measuring method. The Comparative Pathway Analyzer (CPA) calculates and displays the differences between the metabolic reaction contents of several organisms (20). The Roseobacter systems biology database (ROSY) represents an integrated platform for studying the comparative genomics and systems biology of Roseobacter-related species (21). However, even some of the resources for comparative analysis of metabolic pathways of various species focus on a few species only or do not provide a convenient interface for analysis.
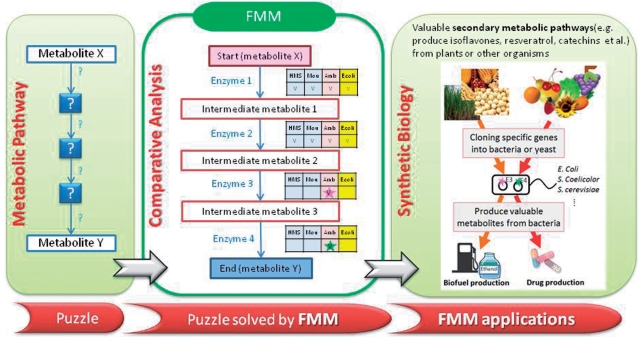
The above considerations motivated the development of a novel and easy resource for use in synthetic biology, which is proposed herein. In this work, a new web server, FMM (From Metabolite to Metabolite), was designed to reconstruct metabolic pathways form one metabolite to another among different species, based mainly on the KEGG database (12–15) and other integrated biological databases. Even though KEGG maps utilized in many metabolic tools, none of them can connect metabolites from different KEGG maps. FMM supports the connection of different KEGG maps. Figure 1 depicts the basic concept of FMM which demonstrates that FMM has numerous applications in synthetic biology and metabolic engineering. For instance, the reconstruction of metabolic pathways to produce valuable metabolites or secondary metabolites in bacteria or yeast is a promising strategy for producing drugs. FMM provides a very convenient way to elucidate which genes from which species should be cloned into those microorganisms based on FMM pathway comparative analysis. Accordingly, FMM is an effective tool in synthetic biology and metabolic engineering, such as applications in drug production and biofuel production.
Figure 1.
A conceptual diagram of FMM.
METHODS
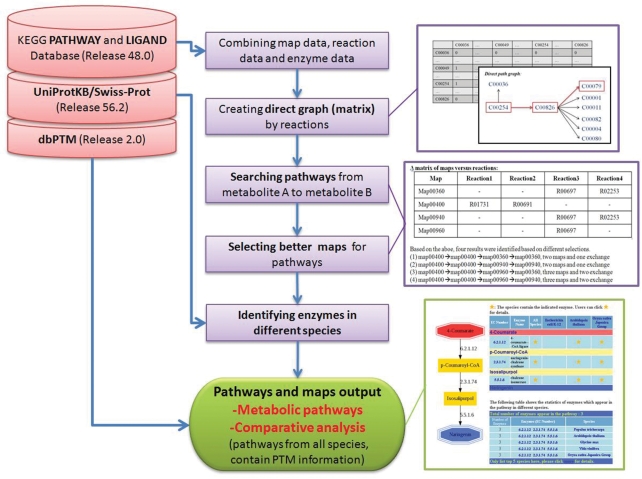
Figure 1 displays metabolic pathways that can be easily reconstructed from two interesting metabolites among various species using the FMM system. Figure 2 presents the system flow of FMM. A reaction matrix was developed to identify numerous reaction processes from one metabolite to another. Enzyme annotations from UniProtKB/Swiss-Prot (22) were employed to identify enzymes from different species in comparative analysis.
Figure 2.
System flow of FMM.
Data collection and integration
FMM reaction definitions, species-specific reactions, reaction maps and enzyme lists were obtained from KEGG/LIGAND and KEGG/PATHWAY database Release 48.0, updated on 1 October 2008 (12–15). Some information (including gene names, Enzyme Commission (EC) numbers, and species-specific enzymes) was supplied from UniProtKB/Swiss-Prot Release 14.0 (22) and NCBI Taxonomy database (23,24). The data in the proposed FMM will be updated regularly. The current metabolites (including ATP, NADH, H2O and CO2) are generally used as carriers for transferring electrons or certain functional groups (including phosphate group, amino group, one carbon unit, and methyl group); they are like the external metabolites that participate in many reactions and are not in pseudo steady state in a sub-network. The connections through current metabolites should be avoided in calculating the path length from one metabolite to another. Hence, the connections through current metabolites and cofactors were deleted manually to make the path length analysis physiologically more meaningful. The current metabolites are displayed in Table S1 (See Supplementary Data). However, some current metabolites are primary metabolites. For example, asparate is not only a current metabolite involved in transamination mechanism (oxaloacetate + NH3 + NADPH = asparate + NADP + H2O), but also a primary metabolite in biosynthesis of amino acids. When the current metabolites are involved in the primary pathway as primary metabolites, such reactions and metabolites are collected in FMM. Therefore, biosynthesis of amino acids can be conducted in FMM.
Posttranslational modification (PTM) is the chemical modification of a protein after its translation. In eukaryote, the PTM of amino acids extends numerous functions of the protein involving protein structure change, such as formation of disulfide bridges and regulation of transcription. However, there is no PTM mechanism after protein translation in prokaryote. Therefore, a functional enzyme that needed PTM in eukaryote was no function when cloning into prokaryote. It is important to know the PTM information before cloning one gene from eukaryote to prokaryote. dbPTM (25) is a database that compiles information on protein post-translational modifications (PTMs), such as the catalytic sites, solvent accessibility of amino-acid residues, protein secondary and tertiary structures, protein domains and protein variations. Protein post-translational modification (PTM) information was extracted from dbPTM (25). The numbers of collected metabolites, enzymes, reactions, species and KEGG maps are 16 832, 2336, 4288, 1491 and 149 in FMM, respectively.
Construction of reaction matrix
Information on reactions and enzymes was obtained from KEGG pathway maps and the equations of each reaction were determined. Therefore, a reaction matrix was constructed based on maps, reactions and enzymes data. For instance, ‘R01731: Oxaloacetate + l-Arogenate <=> l-Aspartate + Prephenate’ is converted into ‘R01731: C00036 + C00826 <=> C00049 + C00254’ (where R01731 is KEGG reaction ID; C00036, C00826, C00049 and C00254 are KEGG compound ID). The reaction R01731 is a reversible reaction which is regarded as two forward reactions in FMM. The direction of one forward reaction is from C00254 to C00036 in the pathway map, the other is from C00036 to C00254. The example illustrate herein is only one forward reaction from C00254 to C00036. Therefore, four reactions are described below:
Each equation corresponds to an edge in a graph; Figure S1 (See Supplementary Data) depicts an example reaction matrix. FMM contains 16884 compounds; and so the size of the reaction matrix is 16884*16884. A direct path graph of the reactions depends on the reaction matrix using the breadth first search (BFS) approach. As shown in Figure S2 (See Supplementary Data), when the direct path graph from compound C00254 to compound C00079 is identified by BFS, the search terminates when the C00079 node is found. Additionally, KEGG MODULE is a new collection of pathway modules, molecular complexes, and other functional units, each represented as a list of KEGG Orthology (KO) identifiers, which were used in pathway reconstruction when the node was a KEGG MODULE.
Reconstruction of metabolic pathway from various KEGG pathway maps
After all possible reaction paths were identified, the number of pathway maps was calculated. Usually, found paths occurred not in only a single pathway map, but in a complicated fashion in several maps. Pathway maps that contain the most paths are selected and the one pathway map that has only one reaction is avoided. A matrix of maps versus reactions was employed to reconstruct metabolic pathway from different KEGG maps. For example, the pathway from C00254 to C00811 passes through several reactions (paths) and maps, as described in Figure S3 (See Supplementary Data). It is better to select fewer maps and fewer map exchanges to reconstruct metabolic pathway. Therefore, based on the above example, ‘map00400 → map00400 → map00360 → map00360’ is much better than ‘map00400 → map00400 → map00960 → map00360’.
COMPARATIVE ANALYSIS
Comparative analysis provided in FMM is useful in synthetic biology. For example, the reconstruction of metabolic pathways to produce valuable metabolites or secondary metabolites in bacteria or yeast is a promising biotechnological strategy. Comparative analysis provides an easy way to elucidate which genes from which species should be cloned into those microorganisms. First, the enzymes identified in the reconstructed pathway were processed to search for orthologous encoding genes from various species. Then, the presence or absence of the pathway in a particular species can be known. Furthermore, comparative tables of several organisms are provided. The species are ranked according to the numbers of enzymes involved in the reconstructed pathway.
WEBSITE USAGE
Web Interface
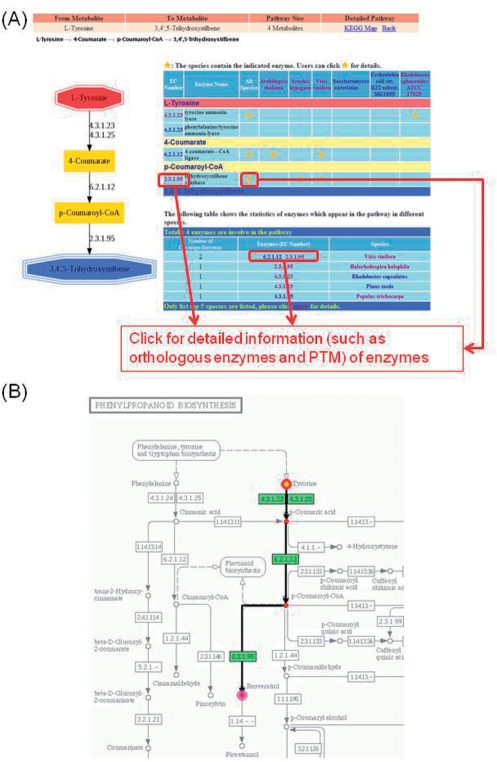
Figure S4 (See Supplementary Data) presents the user-friendly web interface of the FMM. In ‘Start FMM’, two metabolites of interest (keyword or KEGG compound ID) can be input and several species can be selected from four categories (animals, plants, fungi and prokaryotic). The metabolic pathways corresponding to the two input metabolites can be reconstructed, as given in Figure S4A (See Supplementary Data). As to the comparative analysis of metabolic pathways, Figure S4B (See Supplementary Data) includes one species in the ‘major species’ column (of microorganisms commonly used in synthetic biology) and several in the ‘comparative species’ column (of several organisms in common use in the laboratory). The comparative metabolic pathways between various organisms are then generated. The outputs include two views of the metabolic pathway; one is a vertical view (local view); the other highlights the metabolic pathway in the KEGG map (global view), as shown in Figure 3A and B, respectively. The EC number and hyperlinks to detailed information (such as orthologous enzymes and PTM) of the enzymes that are involved in the pathway are supplied when the names of the enzymes and star symbols in the output table and figure are clicked. When the metabolic pathway traverses more than one KEGG map, it can be connected from several KEGG maps using our system, as presented in Figure S5 (See Supplementary Data).
Figure 3.
An example of web interface in FMM. (A) ‘Vertical graph’ and (B) ‘Global graph’ (KEGG map) of the metabolic pathway from tyrosine to resveratrol.
Case studies
Case study I—application in plant secondary metabolites production
Plant-specific flavonoids are important secondary metabolites which are synthesized from 4-coumaroyl coenzyme A (CoA) and derived from the general phenylpropanoid pathway, and three malonyl-CoAs. Numerous plant flavonoid biosynthetic enzymes, which are individually expressed in E. coli as functional enzymes, have been identified (26,27). Furthermore, Watts et al. (28) successfully cloned several critical genes from plants into E. coli and observed the high-level production of flavanone naringenin by feeding exogenous 4-coumarate. In this work, the system of Watts et al.'s was adopted as an example to demonstrate the effectiveness of FMM. When either the genes that can be cloned into E. coli to produce naringenin from 4-coumarate, or the species from which they can be cloned, are unknown, FMM identifies them most easily. 4-Coumarate (C00811) and naringenin (C00509) were inputted into FMM; one E. coli was selected from ‘major species’ and ‘Arabidopsis thaliana’ and ‘Oryza sativa’ were selected from ‘comparative species’ in the ‘Comparative Analysis’ web page. Figure S6 (See Supplementary Data) indicates the effortlessness of understanding how the metabolic pathway from 4-coumarate to naringenin passes through three enzymes (4-coumarate-CoA ligase, chalcone synthase and chalcone isomerase), none of which are present in E. coli. Nevertheless, the results of FMM reveal that all of these genes can be cloned from A. thaliana, Oryza sativa, Populus trichocarpa, Glycine max and Vitis vinifera. Watts et al. (28) cloned 4-coumarate-CoA ligase, chalcone synthase, and chalcone isomerase genes from Arabidopsis into E. coli; the results herein provide information that supports this cloning. FMM is thus a very useful tool in microbial engineering in synthetic biology.
Case study II—application in pharmaceuticals
Plants are well known to generate large amounts of phenylpropanoids which are important sources of pharmaceuticals due to their diverse chemical structures; for example, resveratrol is a chemopreventive anti-cancer agent (29). The inhibition of inflammation, tumor promotion, angiogenesis and metastasis and regulation of cell cycle progression are all pharmaceutical benefits of resveratrol (30). However, the mass-production of resveratrol is expensive because of the extraction processes are costly. However, the superb health effects of resveratrol are motivating scientists to synthesize resveratrol using metabolic engineering methods in bacteria. When synthetic engineering in bacteria, genes from which organisms are cloned into bacteria should be identified. FMM is an effective tool for this purpose. For instance, if E. coli is to be used to produce resveratrol, resveratrol and tyrosine (which is a well known amino acid for the production of plant secondary metabolites) can be input into FMM. The synthesis pathway of resveratrol from tyrosine is easily determined (Figure 3A). Figure 3A reveals that E. coli and yeast cannot synthesize resveratrol unless several critical genes are cloned into their cells from other species. For example, tyrosine-ammonia-lyase can be obtained from R. sphaeroides, 4-coumarate-CoA-lagase can be obtained from V. vinifera or A. thaliana, and trihydroxystilbene synthase can be obtained from A. hypogaea or V. vinifera. The results can be used as candidates for further experimental confirmation. Accordingly, FMM has practical and effective functions in synthetic biology.
Certainly, FMM is also useful in the reconstruction of synthesis (metabolic) pathways. The metabolic pathways of interest can be significantly identified in KEGG maps in FMM. Furthermore, FMM is particularly convenient when the metabolic pathway traverses more than one KEGG map. Figure S5 (See Supplementary Materials) indicates that the metabolic pathway from d-erythrose-4-phosphate to chorismate passes through three KEGG maps. No other server has presented such a convenient interface for identifying metabolic pathway.
SUMMARY
The concept of FMM is illustrated in Figure 1, which demonstrates that metabolic pathways can be easily reconstructed among various species using the FMM system. The results suggest that FMM is helpful in metabolic engineering by reconstructing metabolic pathways for producing some valuable metabolites or secondary metabolites in bacteria or yeast, which approach has potential in drug production. Moreover, FMM can be used to compare the metabolic pathways between numerous species and connect metabolic pathways of different KEGG maps, suggesting that FMM is not only an effective tool for synthetic biology, such as in the production of drugs and biofuels, but also a useful resource for investigation in metabolism.
In order to connect metabolic pathways of different KEGG maps in FMM, the compounds and reactions are restricted by the KEGG database. However, people may argue the incompleteness of FMM database for extracting metabolic pathway, due to several shortcomings of KEGG database, such as incomplete compounds and reactions (31,32). During the prospective works, we will integrate other resources, such as MetaCyc (16) to enhance the information of biochemical reactions in FMM database.
AVAILABILITY
The FMM web server will be continuously maintained and updated. The web server is now freely available at http://FMM.mbc.nctu.edu.tw/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council of the Republic of China (contract numbers: NSC 96-3112-E-009-002, NSC 97-2811-B-009-001 and NSC 97-2627-B-009-007); National Research Program for Genomic Medicine (NRPGM), Taiwan; MOE ATU (Partial). Funding for open access charge: National Science Council of the Republic of China and MOE ATU.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Channon K, Bromley EH, Woolfson DN. Synthetic biology through biomolecular design and engineering. Curr. Opin. Struct. Biol. 2008;18:491–498. doi: 10.1016/j.sbi.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Prather KL, Martin CH. De novo biosynthetic pathways: rational design of microbial chemical factories. Curr. Opin. Biotechnol. 2008;19:468–474. doi: 10.1016/j.copbio.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Park JH, Lee SY. Towards systems metabolic engineering of microorganisms for amino acid production. Curr. Opin. Biotechnol. 2008;19:454–460. doi: 10.1016/j.copbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda M. Amino acid production processes. Adv. Biochem. Eng. Biotechnol. 2003;79:1–35. doi: 10.1007/3-540-45989-8_1. [DOI] [PubMed] [Google Scholar]
- 5.Lee JH, Lee DE, Lee BU, Kim HS. Global analyses of transcriptomes and proteomes of a parent strain and an L-threonine-overproducing mutant strain. J. Bacteriol. 2003;185:5442–5451. doi: 10.1128/JB.185.18.5442-5451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohnishi J, Katahira R, Mitsuhashi S, Kakita S, Ikeda M. A novel gnd mutation leading to increased L-lysine production in Corynebacterium glutamicum. FEMS Microbiol. Lett. 2005;242:265–274. doi: 10.1016/j.femsle.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Ruckert C, Puhler A, Kalinowski J. Genome-wide analysis of the L-methionine biosynthetic pathway in Corynebacterium glutamicum by targeted gene deletion and homologous complementation. J. Biotechnol. 2003;104:213–228. doi: 10.1016/s0168-1656(03)00158-5. [DOI] [PubMed] [Google Scholar]
- 8.Park JH, Lee KH, Kim TY, Lee SY. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc. Natl Acad. Sci. USA. 2007;104:7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA. Proc. Natl Acad. Sci. USA. 2008;105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 11.Keasling JD. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 2008;3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 12.Kanehisa M. Toward pathway engineering: a new database of genetic and molecular pathways. Sci. Technol. Japan. 1996;59:5. [Google Scholar]
- 13.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michal GE. Biological Pathways: An Atlas of Biochemistry and Molecular Biology. New York: Wiley; 1999. [Google Scholar]
- 18.Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, de Ridder D, de Groot MJ, Reinders MJ. Metabolic pathway alignment between species using a comprehensive and flexible similarity measure. BMC Syst. Biol. 2008;2:111. doi: 10.1186/1752-0509-2-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oehm S, Gilbert D, Tauch A, Stoye J, Goesmann A. Comparative Pathway Analyzer – a web server for comparative analysis, clustering and visualization of metabolic networks in multiple organisms. Nucleic Acids Res. 2008;36:W433–437. doi: 10.1093/nar/gkn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pommerenke C, Gabriel I, Bunk B, Munch R, Haddad I, Tielen P, Wagner-Dobler I, Jahn D. ROSY—a flexible and universal database and bioinformatics tool platform for Roseobacter related species. In Silico Biol. 2008;8:177–186. [PubMed] [Google Scholar]
- 22.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 23.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tatusova T, Smith-White B, Ostell J. A collection of plant-specific genomic data and resources at NCBI. Methods Mol. Biol. 2007;406:61–87. doi: 10.1007/978-1-59745-535-0_3. [DOI] [PubMed] [Google Scholar]
- 25.Lee TY, Huang HD, Hung JH, Huang HY, Yang YS, Wang TH. dbPTM: an information repository of protein post-translational modification. Nucleic Acids Res. 2006;34:D622–D627. doi: 10.1093/nar/gkj083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hotze M, Schroder G, Schroder J. Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in Escherichia coli. FEBS Lett. 1995;374:345–350. doi: 10.1016/0014-5793(95)01141-z. [DOI] [PubMed] [Google Scholar]
- 27.Ehlting J, Buttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999;19:9–20. doi: 10.1046/j.1365-313x.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 28.Watts KT, Lee PC, Schmidt-Dannert C. Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. Chembiochem. 2004;5:500–507. doi: 10.1002/cbic.200300783. [DOI] [PubMed] [Google Scholar]
- 29.Yu O, Jez JM. Nature's assembly line: biosynthesis of simple phenylpropanoids and polyketides. Plant J. 2008;54:750–762. doi: 10.1111/j.1365-313X.2008.03436.x. [DOI] [PubMed] [Google Scholar]
- 30.Kundu JK, Surh YJ. Molecular basis of chemoprevention by resveratrol: NF-kappaB and AP-1 as potential targets. Mutat. Res. 2004;555:65–80. doi: 10.1016/j.mrfmmm.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Ginsburg H. Progress in in silico functional genomics: the malaria Metabolic Pathways database. Trends Parasitol. 2006;22:238–240. doi: 10.1016/j.pt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Mombach JCM, Lemke N, da Silva NM, Ferreira RA, Filho EI, Barcellos CK, Ormazabal RJ. Using the FORESTS and KEGG databases to investigate the metabolic network of Eucalyptus. Genet. Mol. Biol. 2005;28:4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.