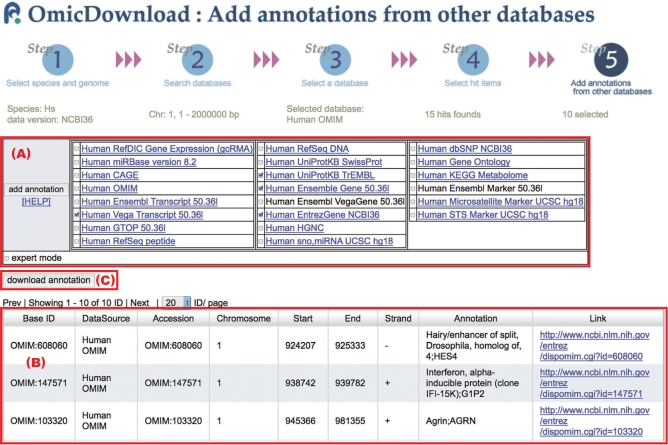
Figure 2.
A screen grab of the data download wizard accessing our OmicDownload server. OmicDownload provides an interactive interface for the users to navigate through a data download procedure consisting the following five steps: (Step 1) selection of species and genome version, (Step 2) search of databases with the user's keyword and chromosomal interval related to the species and the genome version selected in Step 1, (Step 3) selection of a database from the set of databases selected in Step 2, (Step 4) selection of items that satisfy user's keyword and chromosomal interval specified in Step 2 in the database selected in Step 3 and (Step 5) selection of other databases that satisfy Step 1 for each item selected in Step 4, in order to download other items located in the chromosomal interval of the selected item. This figure is a screen grab of Step 5 reached by specifying the following parameters: (Step 1) H. sapiens (Hs) and NCBI36 were selected for species and genome version, respectively, (Step 2) chromosome 1, from 1 Mbp to 2 Mbp was specified as the chromosomal interval, (Step 3) Human OMIM was selected as the database, and 15 records of Human OMIM were found, (Step 4) 10 records were selected by the user from the 15 records obtained in Step 3. In Step 5, four databases related to species Hs and genome version NCBI36 are shown in the database selection table (A). In the annotation table (B), the annotation of the focused Human OMIM record OMIM:608060 is displayed on the top, and other records stored in the selected databases in (A) and located in the chromosomal interval of OMIM:608060 (Chromosome 1, from 924 207 bp to 925 333 bp) are followed. For the other nine records selected in Step 4, related annotation records are displayed as well as for OMIM:608060. Finally, by clicking the download annotation button (C), all annotation data listed in (B) for the 10 records selected in Step 4 are downloaded in a tab separated values (TSV) format.