Abstract
We describe mGene.web, a web service for the genome-wide prediction of protein coding genes from eukaryotic DNA sequences. It offers pre-trained models for the recognition of gene structures including untranslated regions in an increasing number of organisms. With mGene.web, users have the additional possibility to train the system with their own data for other organisms on the push of a button, a functionality that will greatly accelerate the annotation of newly sequenced genomes. The system is built in a highly modular way, such that individual components of the framework, like the promoter prediction tool or the splice site predictor, can be used autonomously. The underlying gene finding system mGene is based on discriminative machine learning techniques and its high accuracy has been demonstrated in an international competition on nematode genomes. mGene.web is available at http://www.mgene.org/web, it is free of charge and can be used for eukaryotic genomes of small to moderate size (several hundred Mbp).
INTRODUCTION
In recent years, the biological community has started to see the dramatic impact of new sequencing technologies on the number of sequenced genomes, and it is expected that this influx of data will continue to escalate in the near future. The demand for efficient, highly automated DNA sequence analysis tools is therefore greater than ever. In particular, we expect that the task of genome annotation will increasingly be performed by individual labs rather than large sequencing centers with dedicated resources and specialized expertise. One of the most important sub-tasks in such an annotation pipeline is the identification of protein coding genes. It requires a computational gene finding system that (i) is highly accurate, (ii) produces genome-wide predictions within a reasonable time, (iii) is easy to use even for researchers with no programming experience and (iv) is applicable to a large variety of newly sequenced organisms. Since computational gene finding has a long tradition in bioinformatics research, there have been constant advancements toward this goal. In particular, the accuracy of computational gene finding systems has steadily been improved, most recently by the introduction of discriminative machine learning techniques (1). However, for this new generation of algorithms, such as mSplicer (14), Craig (2), Conrad (3), Contrast (4), and mGene (Schweikert et al., under review), there is no easy-to-use web application available. To employ these tools, the respective packages have to be downloaded and installed, which in some cases requires substantial programming knowledge as well as the accessibility of sufficient computational power for each user. On the other hand, some of the conventional state-of-the-art gene finders that use generative models, such as Fgenesh (5) and Augustus (6) offer web services, however, without the functionality of training on data provided by the user. Yet, this is essential for accurate annotation of newly sequenced organisms, as models trained on sequences derived from other organisms may produce highly incorrect predictions (7). In 2008, Ter-Hovhannisyan et al. introduced a new ab initio algorithm, GeneMark-ES, that performs unsupervised self-training on anonymous eukaryotic sequences (8). Currently, there is no web service available for GeneMark-ES, in contrast to their self-training prediction program for prokaryotic genomes, GeneMark-S (9). Therefore, to the best of our knowledge, to date no system completely satisfies all the above criteria. We help to close this gap by providing the web service mGene.web that produces highly accurate predictions, is easy to use and allows comfortable training on new data. The high performance of the underlying system mGene (G. Schweikert, under review) has been demonstrated in the international nGASP competition on nematode genomes [Table 1 and (10)]. When considering the average of sensitivity and specificity the evaluated developmental version of mGene exhibited the best prediction performance on nucleotide, exon and transcript level for the ab initio task, and was only slightly worse than Augustus on the gene level. While we have little knowledge on subsequent developments of other participants, we have continued to improve our system after the competition. The fully developed version shows the best performance on all four levels compared with the submitted predictions (Table 1). Additionally, we have verified mGene's high accuracy by biological validation experiments (G. Schweikert, under review).
Table 1.
Comparison of the top-performing gene finding systems in the ab initio setting of the nGASP challenge (10)
| Method | Nucleotide |
Exon |
Transcript |
Gene |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sn | Sp | Sn | Sp | Sn | Sp | Sn | Sp | |||||
| mGene.init | 96.8 | 90.9 | 93.8 | 85.1 | 80.2 | 82.6 | 49.6 | 42.3 | 45.9 | 60.7 | 42.3 | 51.5 |
| mGene.init (dev) | 96.9 | 91.6 | 94.2 | 84.2 | 78.6 | 81.4 | 44.3 | 38.7 | 41.5 | 54.3 | 40.1 | 47.2 |
| Craig | 95.5 | 90.9 | 93.2 | 80.3 | 78.2 | 79.2 | 35.7 | 35.4 | 35.6 | 43.7 | 35.4 | 39.6 |
| Fgenesh | 98.2 | 87.1 | 92.7 | 86.4 | 73.6 | 80.0 | 47.1 | 34.1 | 40.6 | 57.7 | 34.1 | 45.9 |
| Augustus | 97.0 | 89.0 | 93.0 | 86.1 | 72.6 | 79.3 | 52.9 | 28.6 | 40.8 | 64.4 | 34.5 | 49.4 |
Shown are sensitivity (Sn), specificity (Sp) and their average (each in percent) on nucleotide, exon, transcript and gene levels (if several submissions were made for one method, we chose the version with the best gene level average of sensitivity and specificity). The predictions of mGene.init were prepared after the deadline but strictly adhering to the rules and conditions of the nGASP challenge. The result of the best-performing method according to each of the evaluation levels is set in bold face. The evaluation is based on the submitted sets of the participants and performed with our own routine. The numbers slightly deviate from the official nGASP evaluation on the transcript and gene level due to minor differences in the evaluation criteria. These differences, however, do not change the ranking.
Our web server mGene.web provides a convenient interface to mGene for use within the Galaxy framework (11), which also offers handy access to existing genome annotation databases as well as other computational tools. In contrast to most other systems, mGene.web easily allows users to train the prediction model for new genomes. Furthermore, due to the modular structure of mGene.web, the individual sub-tools can be employed independently to predict signals on the DNA, for example transcription starts or splice sites. These predictors are themselves carefully crafted and highly accurate (12,13). However, in combination with Galaxy workflows, the individual units can also easily be replaced by the user when other, possibly more advanced tools for a given sub-task become available. We expect that this particular feature may guide the systematic exploration of further improvements in the complex process of computational gene finding, thereby ultimately leading to more accurate gene predictions.
METHODS: mGene
The precise method for computing accurate gene segmentations employed in mGene is described in detail in (G. Schweikert, under review). Here, we only give a short sketch of the underlying technique, which will help to understand how mGene.web works.
Gene finding can be viewed as a segmentation task, where a piece of genomic DNA has to be properly segmented into genic components, e.g. intergenic regions, UTRs, exons and introns. Such a segmentation is characterized by signals on the genomic DNA that demarcate individual segment boundaries, for instance, splice sites that delineate introns. Each segment type further exhibits a characteristic sequence content, e.g. nucleotide triplets characteristic of coding sequence.
We have taken a two-layered approach and subdivided the problem of gene finding into several sub-tasks that can be solved independently of each other. These tasks include the detection of sequence signals and segment contents. We provide six signal sensors for the detection of transcription start and stop sites, translation initiation and termination sites, as well as donor and acceptor splice sites (12–15). In addition to the signal sensors, we have devised content sensors, which use the sequence of complete segments as input to predict the segment type, i.e. intergenic, 5′-UTR, coding exon, intron or 3′-UTR. Each of these prediction problems is approached with a large-scale implementation of support vector machines using string kernels (as implemented in the Shogun toolbox) that are specifically designed to accurately identify signals and content types [16, and references therein].
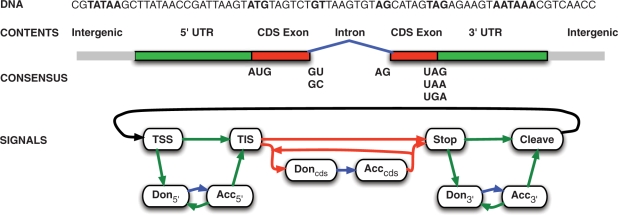
In the second layer, the individual signal and content predictions are reconciled according to a model for gene structures (Figure 1), thereby solving the segmentation task. For this we employ hidden semi-Markov support vector machines (HSM-SVMs) (17), an approach that is conceptually similar to generalized hidden Markov models (gHMMs) (18) in that they are both based on a state-transition model which explicitly parameterizes segment lengths. However, in contrast to generative gHMMs, HSM-SVMs are trained discriminatively. For prediction we use dynamic programming to determine the highest scoring segmentation according to the trained model parameters, thereby predicting accurate gene structures even for new sequences [see for details (15,17) and (G. Schweikert, under review)].
Figure 1.
Simplified gene model underlying mGene: the vertices correspond to recognizable signals on the DNA, i.e. transcription start sites (TSSs), translation initiation sites (TISs), acceptor splice sites (Acc5′, Acccds and Acc3′), donor splice sites (Don5′, Doncds and Don3′), translation termination sites (Stop) and cleavage sites (cleave). The edges correspond to segments associated with the content types for 5′-UTR, coding (CDS) exon, intron, 3′-UTR and intergenic sequences.
WEB SERVER
Essentially, there are two possibilities to use mGene.web for gene predictions: the simplest way is to use the monolithic tools mGenePredict and mGeneTrain. However, for a more flexible use and a more detailed output, one can also use the individual modules together with workflows. In this case, the user has the advantage of accessing intermediate results of the gene prediction task; for example, the outputs of the signal and content sensors can be monitored. Additionally, the progress can be easily observed as the individual tasks are completed. If only sub-tasks should be performed, e.g. the prediction of splice sites, the individual tools can be applied independently. Each option will be described below, and more instructions are provided at http://www.mgene.org/web.
mGene.web monoliths
For organisms that are already in the list of pre-trained models, the simplest way to annotate a genome sequence with mGene.web is to use mGenePredict. This function only requires a FASTA file with the DNA sequence as input and it outputs the GFF3 file containing gene predictions. In case one wishes to annotate a genome for which no suitable pre-trained model exists, mGeneTrain can be used to train a new model. This tool takes a FASTA file with the DNA and a GFF3 file with a set of known genes as input and returns a trained mGene predictor (TmGP) object that can be used together with mGenePredict in order to predict genes on given DNA sequences. The predictor and the predictions can be easily shared with other users via Galaxy's ‘share history’ functionality. We also offer the tool mGeneEval to compare two GFF3 files, presumable one containing the ground truth (an annotation file) and another one containing the predictions. The resulting performance report includes sensitivity and specificity values on nucleotide, exon, transcript and gene level as well as for several signals (like transcription start and stop site) and enables a convenient quality control. For instructions and example runs go to mGene.web → Examples and Instructions and http://www.mgene.org/web/examples.
mGene.web workflows
Recently, Galaxy has started to support workflows that allow pre-defining the order in which tools are applied to data files (including original inputs or interim results) in order to achieve a certain goal. These workflows can be edited by, and shared among users. Workflows can help to simplify the relatively complex process of creating a gene finding system which involves many different steps. In order to run a workflow, one only needs to specify a few input arguments. As a result one obtains, for instance, the gene predictions as well as intermediate results allowing detailed inspection. Analogous to the simple black-box versions, we provide two workflows (The monolithic workflows described above are equivalent to the modular versions. They show less internal details and are slightly more efficient): mGenePredict and mGeneTrain. For more information go to mGene.web workflows → Examples and Instructions or http://www.mgene.org/web/workflows. There, you will also find instructions on how to built new workflows, in which individual tools are replaced by other, potentially improved ones.
mGene.web modules
The web service currently provides 14 core modules. They can be grouped into four groups: data preparation; signal training and prediction; content training and prediction; and gene structure training and prediction. Each tool requires a set of inputs and provides at least one output. They are managed by the Galaxy system according to their data types. We use the following data types for data exchange between the modules: FASTA (genomic DNA), GFF3 (genome annotation and gene prediction), SPF (signal prediction format) and CPF (content prediction format). Files in these formats can either be provided by uploading or by running one of the tools. Additionally, we have some data formats representing internal data structures, including the Genome Information Object (GIO) to describe a genome-wide sequence with all its contigs or chromosomes, the Annotation Gene Structure (AGS), as an efficient internal representation of genes, the Trained Signal Predictor (TSP), the Trained Content Predictor (TCP) and the Trained Gene Predictor (TGP) that include all learnt parameters necessary to predict a given signal, content or gene structure, respectively. Eventually, the trained mGene predictor (TmGP) contains all above-mentioned parameters and can be used to predict genes from scratch when given a DNA sequence. (Note that the TGP object contains the parameters learnt during training layer 2 only, while the TmGP contains all parameters necessary to predict genes.) In the following, the individual modules are explained in more detail (see also mGene.web modules → Examples and Instructions and http://www.mgene.org/web/modules.
Data preparation
GenomeTool needs a file in FASTA format containing genomic sequences as input that allows it to create a genome object, stored in a GIO, to be used by other mGene modules. Additionally, one may create a GIO from an internal database of more than 50 genomes.
GFF2Anno reads an annotation file in GFF3 or GTF format and uses it to generate an AGS. It provides predefined settings for commonly used GFF-encoding conventions, but also permits specific settings needed for less common encoding choices.
Signal prediction
Anno2SignalLabel uses an AGS to collect labeled genomic positions for the selected genomic signal. Possible signals include transcription start and stop sites, translation initiation and termination sites, as well as donor and acceptor splice sites. It uses the regions covered by annotated features to generate negative examples at all consensus positions unless they were annotated as true sites. The output is a file in SPF providing chromosome/contig name, position, strand and the label of the example.
SignalTrain trains a signal predictor using SVMs with pre-selected kernels for each signal. Input is a GIO and an SPF file with labeled genomic positions. The output is a TSP that can be used with SignalPredict to perform predictions on genomic sequences.
SignalPredict uses a GIO and TSP to predict signals on the given DNA sequences. The output is given in SPF.
SignalEval takes a label file and a prediction file (both SPF files) as input and computes several accuracy measures for the predictions, including the areas under the Receiver Operator Curve (ROC) and the Precision Recall Curve (PRC). This tool is useful for prediction quality monitoring.
Content prediction
Anno2ContentLabel collects labeled genomic segments for the selected content types, analogous to Anno2SignalLabel. Possible content types include 5′-UTR, exonic, intronic, 3′-UTR and intergenic. Any segment included that is not of the specified type is used as a negative example. The output is a file in CPF providing chromosome/contig name, start position, end position, strand and the label of the example.
ContentTrain is analogous to SignalTrain, with a GIO and an SPF file as inputs and a TCP object as output.
ContentPredict is analogous to SignalPredict, with a GIO and a TCP as input and an SPF file as output.
ContentEval, analogous to SignalEval, takes a CPF and SPF file as input and performs the performance evaluation.
Gene prediction
GeneTrain trains the second layer of mGene.web. Based on the GIO, genome-wide predictions for all relevant signals and content types, and a set of annotated genes, GeneTrain learns to predict gene structures from genomic DNA. The output is an internal data structure containing the TGP that can be used with GenePredict to predict genes.
GenePredict uses the TGP (either from the current history or from a list of pre-trained predictors) as well as genome-wide signal and content predictions to predict genes from the provided DNA sequences. The output is provided as a GFF3 file.
GeneEval takes two GFF3 files, one containing an annotation, the other the genome-wide gene predictions, and evaluates the prediction performance by comparing the two annotations. Note that the annotated genes should be distinct from the annotated genes used for training, otherwise the training error will be reported. Evaluation criteria include sensitivity and specificity on nucleotide, exon and gene levels [(10) and (G. Schweikert, under review) for further details].
ComposeMGenePredictor bundles all necessary trained signal, content and gene predictor objects into a TmGP that can be used with mGenePredict to predict genes.
COMPUTING TIME AND RESOURCES
The web service is running on a cluster with 84 AMD Opteron CPUs (2.2 GHz) with 8 GB of RAM per four CPUs which is shared with other web services. Training and prediction is split into several parallel sub-tasks in order to reduce the waiting time for users. Depending on the load of the cluster, whole-genome prediction of all six signals for the Caenorhabditis elegans genome (100 Mbp) takes ∼24 h (about ≈500 kbp/min), the prediction of the five content types also takes about 2 h (about ≈1 Mbp/min), and gene prediction using these signals takes ∼6 h (∼300 kbp/min). The time for training the predictors strongly depends on the amount of available training data. While signal or content sensor training can typically be completed for a genome like that of C. elegans within a few hours, training the second layer of mGene.web may take 48 h (depending on the size of the training set).
For well-annotated genomes, extensive annotations exist. In these cases, we have to limit the amount of data used for training the system (due to the relatively high memory and computing time demand). In these cases, we sub-sample the data and may therefore obtain sub-optimal results compared with including all data. mGene.web currently works best for a few well-annotated regions of a genome with chromosomes/contigs of size <20 Mb, including at least a few hundred and at most a few thousand genes. Currently, the speed of the system is severely influenced by the number of chromosomes/contigs, as each one is processed separately. Moreover, there are a few built-in settings that are more suitable for organisms with compact genomes, for instance, that introns have to be shorter than 20 kb. We intend to make these options configurable in the near future.
DISCUSSION AND OUTLOOK
To the best of our knowledge, with mGene.web we currently provide the only web service that offers both prediction and complete training of a gene finding system for eukaryotic genomes. The underlying Galaxy framework greatly facilitates the data acquisition and supports workflows that appear very useful for the rather complex processes needed for genome annotations. In particular, it follows simple and standardized interfaces for data processing, training, prediction and evaluation. Advanced users may take advantage of the modular nature of mGene.web and compose their own Galaxy workflows. This should facilitate the incorporation of new developments and improvements of individual modules and will therefore be of great value for the advancement of gene finding systems.
In addition to the existing features, we will soon extend our web service in several respects. First, we will allow the utilization of additional data sources, such as EST or whole-genome alignments, as implemented in mGene.seq and mGene.multi (G. Schweikert, under review). Another line of improvements will aim at providing more advanced tools for the alignment of sequences of mRNA and orthologous proteins, thereby generating initial gene annotations for training. This will be an important step toward our next goal, to offer accurate gene prediction services for new genomes requiring no or considerably fewer known genes for training an accurate predictor.
FUNDING
Funding for open access charge: Friedrich Miescher Laboratory of the Max Planck Society.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We gratefully acknowledge the great support we received from the Galaxy development team at Penn State University, in particular James Taylor. Moreover, we would like to thank Fabio De Bona for developing an Octave interface to the MOSEK optimization toolbox as well as Peter Niermann and Zhiqin Huang for integrating new genomes and testing the system.
Footnotes
Present address: Cheng Soon Ong, Department of Computer Science, ETH, Zürich, Switzerland.
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Brent MR. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat. Rev. Genet. 2008;9:62–73. doi: 10.1038/nrg2220. [DOI] [PubMed] [Google Scholar]
- 2.Bernal A, Crammer K, Hatzigeorgiou A, Pereira F. Global discriminative learning for higher-accuracy computational gene prediction. PLoS Comput. Biol. 2007;3:e54. doi: 10.1371/journal.pcbi.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeCaprio D, Vinson JP, Pearson MD, Montgomery P, Doherty M, Galagan JE. Conrad: gene prediction using conditional random fields. Genome Res. 2007;17:1389–1398. doi: 10.1101/gr.6558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross SS, Brent MR. Using multiple alignments to improve gene prediction. Comput. Biol. 2006;13:379–393. doi: 10.1089/cmb.2006.13.379. [DOI] [PubMed] [Google Scholar]
- 5.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanke M, Schoffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinf. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korf I. Gene finding in novel genomes. BMC Bioinf. 2004;5:S7. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;12:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coghlan A, Fiedler T, McKay S, Flicek P, Harris T, Blasiar D, The nGASP Consortium, Stein L. nGASP: the nematode genome annotation assessment project. BMC Bioinf. 2008;9:549. doi: 10.1186/1471-2105-9-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giardine B, Riemer C, Hardison R, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonnenburg S, Zien A, Rätsch G. Accurate recognition of transcription starts in human. Bioinf. 2006;22:e472. doi: 10.1093/bioinformatics/btl250. [DOI] [PubMed] [Google Scholar]
- 13.Sonnenburg S, Schweikert G, Philips P, Behr J, Rätsch G. Accurate splice site prediction using support vector machines. BMC Bioinf. 2007;8(Suppl 10):S7. doi: 10.1186/1471-2105-8-S10-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rätsch G, Sonnenburg S, Srinivasan J, Witte H, Müller K.-R, Sommer R, Schölkopf B. Improving the C. elegans genome annotation using machine learning. PLoS Com. Biol. 2007;3:e20. doi: 10.1371/journal.pcbi.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonnenburg S, Rätsch G, Rieck K. Large-scale learning with string kernels. In: Bottou L, Chapelle O, DeCoste D, Weston J, editors. Large-Scale Kernel Machines, Ch. 4. Cambridge, MA: MIT Press; 2007. pp. 73–104. [Google Scholar]
- 16.Ben-Hur A, Ong C, Sonnenburg S, Schölkopf B, Rätsch G. Support vector machines and kernels for computational biology. PLoS Com. Biol. 2008;4:e1000173. doi: 10.1371/journal.pcbi.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rätsch G, Sonnenburg S. Large-scale hidden semi-Markov SVMs. In: Schölkopf B, Platt J, Hoffman T, editors. Advances in Neural Information Processing Systems (NIPS'06). Vol. 19. Cambridge, MA: MIT Press; 2007. pp. 1161–1168. [Google Scholar]
- 18.Kulp D, Haussler D, Reese M, Eeckman F. A generalized hidden Markov model for the recognition of human genes in DNA. ISMB. 1996;4:134–141. [PubMed] [Google Scholar]