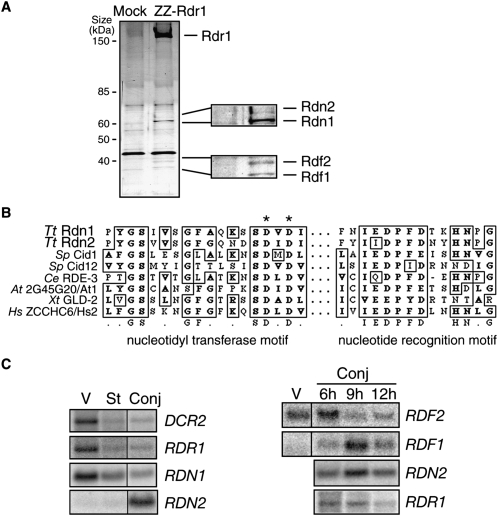
FIGURE 1.
Rdr1-associated RDRC subunits are two pairs of related proteins with differential mRNA expression profiles. (A) Extracts from starved cells with no tagged protein (Mock) or with ZZ-Rdr1 were used for affinity purification of RDRC. The four proteins recovered specifically in association with Rdr1 are labeled in the enlargements, with assignments based on mass spectrometry, protein tagging, and genetic depletion assays. (B) An alignment of active site residues in Rdn1, Rdn2, and other noncanonical ribonucleotidyl transferases is shown, with consensus underneath. Identical and similar residues are boxed. Asterisks denote residues mutated to alanine to render this class of enzyme catalytically inactive. Nucleotidyl transferase and nucleotide recognition motifs are indicated as previously described (Martin and Keller 2007). Tt, Tetrahymena thermophila; Sp, Schizosaccharomyces pombe; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana; Xt, Xenopus tropicalis; Hs, Homo sapiens. (C) Northern blots for mRNA expression were performed using total RNA isolated from cells in vegetative growth (V), starvation (St), or conjugation (Conj). Time points after the initiation of conjugation are noted in hours (h); where not noted, the 10-h time point of conjugation was used (see Fig. 3B for conjugation stages).