Abstract
Small RNAs play important roles in the establishment and maintenance of heterochromatin structures. We show the presence of telomere specific small RNAs (tel-sRNAs) in mouse embryonic stem cells that are ∼24 nucleotides in length, Dicer-independent, and 2′-O-methylated at the 3′ terminus. The tel-sRNAs are asymmetric with specificity toward telomere G-rich strand, and evolutionarily conserved from protozoan to mammalian cells. Furthermore, tel-sRNAs are up-regulated in cells that carry null mutation of H3K4 methyltransferase MLL (Mll(−/−)) and down-regulated in cells that carry null mutations of histone H3K9 methyltransferase SUV39H (Suv39h1/h2(−/−)), suggesting that they are subject to epigenetic regulation. These results support that tel-sRNAs are heterochromatin associated pi-like small RNAs.
Keywords: heterochromatin assembly, histone methylation, piRNA
INTRODUCTION
In the past decade, several classes of small RNAs have been characterized in mammals including small interfering RNAs (siRNAs), microRNAs (miRNAs), and Piwi-interacting RNAs (piRNAs) (Birchler and Kavi 2008). siRNAs and miRNAs are 20–22 nucleotide (nt) small RNAs in association with the Argonaute (Ago) subfamily of Ago-Piwi proteins. They are derived from double-stranded RNA (dsRNA) precursors in a Dicer-dependent pathway and widely distributed in germ line and somatic cells (Stadler and Ruohola-Baker 2008). In contrast, piRNAs are longer (24–30 nt), associated with Piwi-subfamily proteins, and derived from single stranded RNAs (ssRNA) by a Dicer-independent mechanism (Aravin et al. 2007a; Klattenhoff and Theurkauf 2008). piRNAs in mammals have only been reported in the male germ line (Aravin et al. 2006; Girard et al. 2006; Lau et al. 2006; Aravin et al. 2007b). These small RNAs have been demonstrated to play various important roles in modulating gene expression, regulating stem cell self-renewal, providing defense against invading DNA, and establishing chromosomal architecture.
Extensive studies, mostly in fission yeast, demonstrate that small RNA-dependent processes are central for the formation of constitutive heterochromatin (Moazed et al. 2006; Grewal and Elgin 2007). In this pathway, siRNAs generated by a Dicer homolog (Dcr1) associate with an Argonaut homolog (Ago1) and help to recruit histone-modifying activities to chromatin regions with homologous sequences. In particular, histone methyltransferase Clr4, a Drosophila Su (var) 39 homolog, methylates H3K9, leading eventually to the formation of condensed heterochromatin structures (Nakayama et al. 2001; Verdel et al. 2004; Zhang et al. 2008). A similar mechanism for heterochromatin assembly in mammalian cells was suggested, but has yet to be firmly established. It remains controversial whether there are Dicer-dependent small RNAs in association with repeat sequences and heterochromatin, despite several recent large-scale cloning and sequencing efforts to delineate the small RNA profiles in both wild-type and Dicer (−/−) mouse ES cells (Kanellopoulou et al. 2005; Murchison et al. 2005; Calabrese et al. 2007).
An important aspect of the regulation and functioning of telomeres is their chromatin structures (Blasco 2007b). In mammalian cells, telomeres are assembled into nucleosomes. Both telomeric and subtelomeric chromatin are enriched in heterochromatic histone modifications, such as trimethylation of H3K9 and H4K20 (Blasco, 2007a), while there are a low abundance of euchromatic marks, such as acetylated H3 and H4. Increasing evidence indicates that there is a functional association between heterochromatin formation and telomere-length homeostasis. For example, mutations in the histone methyltransferases, such as Suv39h1/h2 or Suv4-20, lead to drastically reduced H3K9me3 or H4K20me3, respectively, which are concomitant with loss of telomere length control and genome instability (Garcia-Cao et al. 2004; Benetti et al. 2007b). Conversely, mutations in telomere-associated proteins and telomerase (TERT) can bring about changes to the telomeric chromatin structures and/or epigenetic status. Knocking out TERT or components of the shelterin complex leads to reduced H3K9me3 at the telomere and subtelomere regions (Benetti et al. 2007a). The mechanism underlying the telomeric heterochromatin assembly remains poorly understood, and its elucidation promises to provide insights into telomere-mediated genome instability, as well as replicative senescence.
Small RNAs have been implicated in heterochromatin assembly in subtelomeric regions of fission yeast (Kanoh et al. 2005). Transcription repression at the chromosomal ends, also referred to as telomere position effect (TPE), requires the H3K9-specific histone methyltransferase Clr4, as well as components of the RNAi machinery (Kanoh et al. 2005). However, the short telomeres of yeast are not assembled into nucleosomes. Similar functions of small RNAs in telomere heterochromatin assembly have not been reported in mammalian cells. Recently, noncoding RNA transcripts (TERRA or Tel-RNA) up to several kilobases long are detected at telomere and subtelomere regions in mammalian cells (Azzalin et al. 2007; Luke et al. 2008; Schoeftner and Blasco 2008). The levels of these long telomere transcripts are dependent on RNA polymerase II, and are also regulated by development status, telomere length as well as chromatin structures at telomeres (Azzalin et al. 2007; Schoeftner and Blasco 2008). As noncoding RNA transcripts are often precursors to small RNAs and are required for establishing RNAi-dependent heterochromatin assembly in a wide range of eukaryotic systems (Grewal and Elgin 2007), we decided to test whether telomere specific small RNAs are present in mammalian cells, and if so, whether these small RNAs are involved in the epigenetic regulation of telomeres.
RESULTS AND DISCUSSION
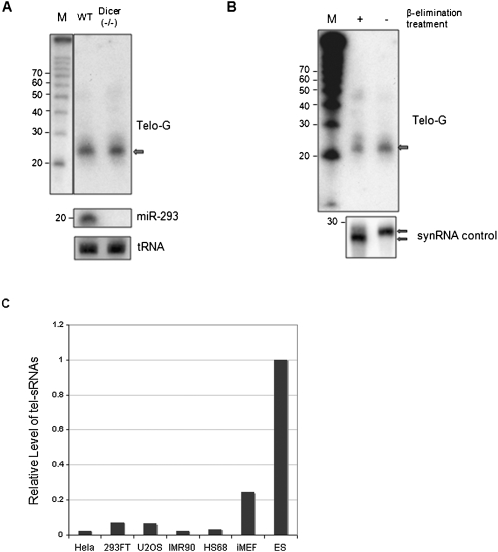
In order to test for the presence of telomere-specific small RNAs in mammalian cells, we purified small RNA-enriched RNA samples from mouse embryonic stem cells (ES). These RNAs were then treated with DNase I, blotted, and probed with 32P end-labeled oligos (TTAGGG)3 or (CCCTAA)3. Telomeric-specific small RNAs (tel-sRNAs) corresponding to the G-strand were detected, with an estimated length of 24 nt (Fig. 1A), which is longer than small interfering RNAs (siRNAs) and micro-RNAs (miRNAs). We could not detect any C-strand specific tel-sRNAs, indicating that the generation of tel-sRNAs is asymmetric and G-strand-specific (UUAGGG)—similar to the recently reported telomere-associated long RNA transcripts (TERRA/TelRNA). In order to further confirm that there are telomere-repeat-containing small RNAs in vivo, we specifically amplified the tel-sRNAs by PCR (see Materials and Methods), cloned the PCR products into the TA vector, and sequenced 20 individual clones using a primer specific for TA vector. From 15 sequences obtained from 20 clones, 87% of the clones contained the partial telomere repeat sequences. Among them, 33% of the clones contain asequence of TAGGG, TAGG, or TAG between the primer, (TAGGGT)3, and poly A sequences of the PCR products. Fifty-four percent clones have either thymidine (T) or adenosine (A) at the junction of telomere repeats and poly(A) sequence. This result suggests that tel-sRNAs are heterogeneous populations. The narrow size distribution of the signals detected by Northern blot analysis (Fig. 1A,B) argues against it being an artifact of random RNA degradation. Instead, the size and asymmetric nature of tel-sRNAs are suggestive of Piwi-interacting small RNAs (pi-RNAs) recently found in germ-line stem cells in Drosophila, zebrafish, and mammals. In order to decide whether tel-sRNAs are pi-like small RNAs or siRNAs, we tested the level of tel-sRNAs in Dicer(−/−) cells. As shown in Figure 1A, comparable levels of tel-sRNAs in wild type and Dicer(−/−) embryonic stem cells were detected by Northern blot analysis. In contrast, miR-293, a miRNA expressed in ES cells, was completely abolished in Dicer(−/−) cells (Fig. 1A, middle panel). Northern blot using the tRNA-Ile-AAT probe was included as a loading control (Fig. 1A, bottom panel). In addition to Dicer independence, another feature that is unique to piRNAs is the 2′-O-methylation at the 3′ terminus. This modification blocks periodate oxidation (NaIO4) and β-elimination of the piRNAs (Gunawardane et al. 2007). To test whether tel-sRNAs are resistant to β-elimination reaction, the small RNAs from mouse ES cells were subjected to oxidation and β-elimination treatment. As a control, we treated a synthetic RNA probe that is not 2′-O-methylated in parallel. As expected, the unblocked control small RNA migrates faster on the denatured gel after treatment (Fig. 1B, bottom panel). In contrast, the mobility of tel-sRNAs on the gel remains the same after treatment, suggesting that they are protected at the 3′ terminus, a feature unique to pi-RNAs (Gunawardane et al. 2007). The narrow size distribution, as well as the 3′ block by 2′-O-methylation strongly argues that the tel-sRNAs are not the abortive transcripts or the degradation products of long telomere transcripts TERRA/Tel-RNAs. Instead, tel-sRNAs are most likely pi-like small RNAs. It is worth noting that unlike the germ-line specific pi-RNAs in mammals, tel-sRNAs were detected in ES cells (Fig. 1A), as well as other somatic cells like mouse embryonic fibroblast (MEF) cells, albeit at much lower abundance (Fig. 1C).
FIGURE 1.
Analysis of telomeric small RNA transcripts in mouse embryonic stem cells. (A) Total RNA was size fractionated using a PureLink miRNA isolation kit. Samples of small RNA molecules from wild-type and Dicer(−/−) cells were analyzed by Northern blot after DNase I treatment. A32P-end labeled (CCCTAA)3 oligonucleotide probe was used to detect small-sized telomeric transcripts. For sizing control, the radio labeled RNA decade marker (Ambion) is shown on the left. The blot was reprobed with miR-293 (middle panel) and tRNA-Ile-ATT, which serves as a loading control (bottom panel). (B) The small RNAs treated with and without sodium periodate and β-elimination were separated on a 15% gel. The 32P-end labeled (CCCTAA)3 oligonucleotide probe was used to detect small-sized telomeric transcripts. RNA marker is shown on the left. As a control, a synthetic RNA oligo (with free 2′OH) treated with or without sodium periodate and β-elimination were 5′ end labeled with 32P and separated on a 12% gel. (C) Quantitative PCR for tel-sRNAs in various cell lines as indicated on the bottom.
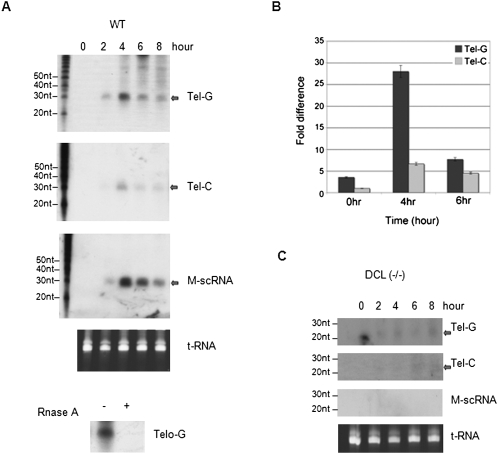
To test whether the presence of tel-sRNAs is evolutionarily conserved, we performed small RNA Northern blot analyses in the ciliate Tetrahymena thermophila, an early branching eukaryote. A particular class of small RNAs, scan RNAs (scnRNAs), are abundantly present in Tetrahymena during conjugation, the sexual phase of the Tetrahymena life cycle (Mochizuki and Gorovsky 2004). Total RNAs were extracted at different conjugation time points, blotted, and hybridized with telomere specific probes (TGGGGT)3 or (ACCCCA)3. We were able to detect tel-sRNAs, which are RNase A sensitive, homologous to both the G- and C-strands (Fig. 2A). However, the level of G-strand is much higher than that of the C-strand, which can only be detected after prolonged exposure time. This result is consistent with the observation in mammalian cells that tel-sRNAs has strong G-strand bias. We further confirmed this by small RNA quantitative PCR experiments (Fig. 2B). Tel-sRNAs in Tetrahymena were about 28 nt in length and their levels peaked at 4 h after the initiation of conjugation, similar to other well-characterized scnRNAs, such as M-related scnRNAs (Fig. 2A; Liu et al. 2004; Malone et al. 2005). ScnRNAs in Tetrahymena share many features with piRNAs in metazoans. These include association with Piwi-clade, but not Ago-clade of the Piwi-Ago family proteins (Mochizuki et al. 2002); increased length; and enrichment in germ line, especially meiotic cells. One notable difference between scnRNAs and piRNAs is the observed Dicer dependence of the scnRNA biogenesis (Mochizuki and Gorovsky 2004). In the absence of DCL1 (Dicer-like 1), which is essential for processing scnRNA, tel-sRNA levels were dramatically reduced. Interestingly, increased levels of shorter telomere-specific small RNAs (23–24 nt) were detected in Dcl1(−/−) cells (Fig. 2C, top panel). The abnormal accumulation of the 23–24 nt small RNAs in conjugating Dcl1(−/−) cells has been previously reported (Malone et al. 2005; Mochizuki and Gorovsky 2005), but until now their identity was obscure, as they do not cross-react with well-established scnRNAs, such as M-related scnRNAs (Fig. 2C, bottom panel). The 23–24 nt small RNAs are likely processed by other Dicer-homologs (DCR1 and DCR2), as a compensation mechanism in the absence of DCL1. The results underscore the complexity of the biogenesis pathway of tel-sRNAs in Tetrahymena. Nonetheless, the telomere associated small RNAs are evolutionarily conserved.
FIGURE 2.
Tel-sRNA generation is correlated with genome reorganization and heterochromatin formation in Tetrahymena thermophila. (A) Northern blot analyses of RNAs collected at different times of conjugation (shown on top) from wild-type cells. For RNase treatment, RNAs extracted at 4 h of conjugation were used. (B) Quantitative RT-PCR of tel-sRNAs using RNA samples in A. Signals were presented as fold differences relative to Tel-C signal in 0 h of conjugation, which was arbitrarily set as 1. (C) Northern blot analyses of RNAs collected at different time of conjugation (shown on top) from or Dcl1(−/−) cells. In both A and C, the blots were hybridized with (ACCCCA)3, (TGGGGT)3, or M-scnRNA probes, respectively. The transcripts were indicated on the right. Ethidium bromide stained PAGE gel was shown at bottom as the loading control.
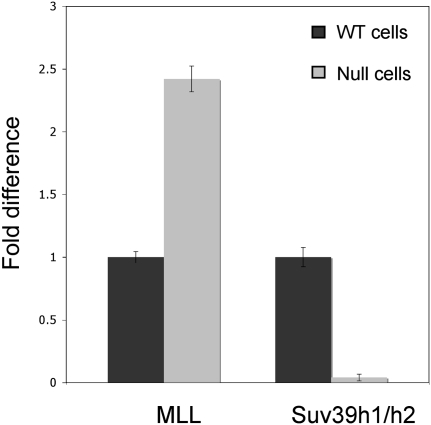
The evolutionary conservation of tel-sRNAs suggests that they may play important roles in telomere structure and/or functions. A positive feedback mechanism involving siRNAs and repressive histone modifications, especially H3K9 methylation, has been well established for heterochromatin assembly in pericentromeric regions (Grewal and Elgin 2007). A similar mechanism has been suspected, but never demonstrated, for telomeric heterochromatin. Here we tested whether tel-sRNA levels are regulated by different chromatin modifying activities in mammalian cells. We focused on Mll, which mediates H3K4 methylation (Milne et al. 2002; Dou et al. 2005), and Suv39h1/h2, which deposits H3K9 methylation (Rea et al. 2000). These two antagonistic histone methylation events are often used to define the euchromatin and heterochromatin regions, respectively. We carried out the experiments using Mll(−/−) (McMahon et al. 2007) and Suv39h1/h2(−/−) MEF cells (Peters et al. 2001), and their corresponding wild-type controls. The level of tel-sRNA is significantly lower in MEF cells compared to ES cells (Fig. 1C). However, we were able to detect and quantify tel-sRNAs using the previously described small RNA specific quantitative PCR analysis. We found that G-strand tel-sRNAs increase twofold in Mll(−/−) cells, while they decrease by more than tenfold in Suv39h1/h2(−/−) cells (Fig. 3). These results support that tel-sRNAs in mammals are regulated by epigenetic mechanisms. It is worth noting that the effect we observed in Suv39h1/h2(−/−) cells is opposite to the previously reported telomere long transcripts TelRNA or TERRA, which are slightly increased in Suv39h1/h2(−/−) cells.
FIGURE 3.
Tel-sRNA level is regulated by chromatin modifying activities. Quantitative RT-PCR was performed using RNA extracted from wild type and either Mll(−/−) or Suvh1/h2(−/−) knockout MEF cells. Knockout genotypes are indicated at bottom. Signals in knockout cells were presented as fold differences relative to control MEF cells. All RNA samples were treated with DNase I prior to reverse transcription.
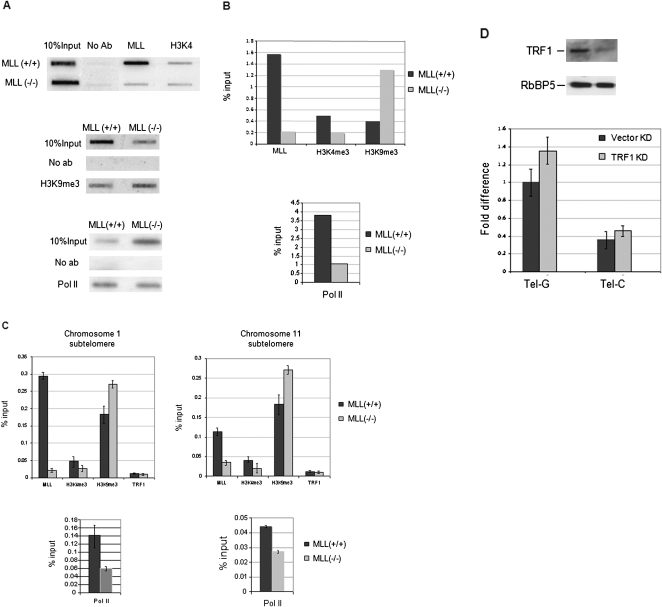
Increased tel-sRNAs in Mll(−/−) cells is somewhat surprising, as Mll is usually associated with transcriptionally active genes in euchromatin. To test whether Mll regulate tel-sRNAs by playing a direct role in telomere chromatin dynamics, we examined the Mll binding by chromatin immunoprecipitation (ChIP) assays. We found that Mll physically associated with both telomere (Fig. 4A,B) and subtelomere regions (Fig. 4C). Furthermore, we found the presence of both the heterochromatin mark H3K9me3 and the euchromatin mark H3K4me3, although H3K4me3 was at much lower levels than H3K9me3. In the absence of Mll, the level of H3K4me3 at telomeres decreased and there was a dramatic increase in H3K9me3. We also observed reduced binding of RNA polymerase II at both telomeres and subtelomere regions in Mll(−/−) cells (Fig. 4A–C). Both the increased H3K9 methylation and the decreased RNA polymerase II binding in Mll(−/−) cells reflect a shift toward more heterochromatic structures at telomeres in the absence of Mll. This suggests that there is dynamic equilibrium between euchromatin and heterochromatin at mammalian telomeres. Interestingly, the increased heterochromatic feature is parallel to the increased levels of tel-sRNAs (Fig. 3), lending further support to the mechanistic connection between telomeric heterochromatin and tel-sRNAs. Modulation of the extent of heterochromatin formation by H3K4 methylation may be a common theme. This is supported by the observation that mutation of Drosophila Su (var) 3-3, a homolog of mammalian H3K4 demethylase LSD1, was shown to impair the full expansion of H3K9me2 marks at the chromosomal end (Rudolph et al. 2007).
FIGURE 4.
MLL regulates chromatin equilibriums at both subtelomeres and telomeres. (A) Slot-blot analysis of ChIP DNA using indicated antibodies. The telomere specific probe was used for the hybridization. (B) Quantification of slot-blot results in A. Signals in ChIP DNA was normalized against 10% input using Image J software. (C) MLL regulates chromatin structures at subtelomere regions. ChIP experiments in Mll(+/+) and Mll(−/−) using the indicated antibodies. ChIP results are quantified using the primer sets specific for subtelomere region of chromosome 1 (left) or chromosome 11 (right). Signals are normalized as a percent input. Standard deviations are indicated by error bars. (D) Top, immunoblot was used to show the reduced levels of TRF1 in stable TRF1 knockdown cells. Immunoblot of RbBP5 protein, a component of Mll complex, was used as a loading control. Bottom, Quantitative RT-PCR for tel-sRNA in TRF1 knockdown cells.
In addition to chromatin components, telomere structure is also regulated by the shelterin complex, which forms a dynamic terminal structure that caps the natural ends of linear chromosomes (Cech 2004; de Lange 2005). Therefore, we examined the role of the shelterin complex in regulating tel-sRNA levels. To this end, we stably knocked down TRF1, a key component of the shelterin complex, in MEF cells (Fig. 4D, top panel). TRF1 protein levels were greatly diminished as indicated by the Western blot analysis (Fig. 4D, top panel). However, the level of tel-sRNAs was not significantly affected (Fig. 4D, bottom). This is consistent with our observations that there are only slight changes of chromatin modifications at both telomere and subtelomere regions in our TRF1 knockdown cells (data not shown).
Direct roles of small RNAs in the propagation of heterochromatin have been established in several eukaryotic systems, including Tetrahymena, fission yeast, and Drosophila (Moazed et al. 2006; Zofall and Grewal 2006; Liu et al. 2007). However, the case remains unresolved in mammalian cells, in large part due to the difficulty of detecting small RNAs homologous to heterochromatin regions, especially in somatic cells. Here we report the presence of telomere repeat (UUAGGG) containing small RNAs in mammalian cells and in a lower eukaryote, Tetrahymena thermophila. The tel-sRNAs in mammalian cells have the following characteristics: (1) asymmetric with specificity for the G-strand; (2) Dicer independent; (3) 2′-O-methylated at the 3′ end; (4) present in embryonic stem cells, as well as somatic MEF cells; and (5) subject to regulations by histone modifying enzymes, such as MLL and Suv39 h1/h2. All these strongly suggest that tel-sRNAs are pi-like RNAs associated with the telomeric heterochromatin.
Previously reported piRNAs in mammalian cells are restricted to the male germ line, where Piwi family proteins (i.e., MIWI, MILI, and MIWI2) are specifically expressed (Aravin et al. 2007a). The finding of pi-like tel-sRNAs in mouse somatic cells raises the question whether the Piwi-related slicer activity is present in cells other than male germ line cells. Expression of MILI in prenatal ovaries was reported (Kuramochi-Miyagawa et al. 2001), suggesting that not all Piwis in mice are exclusively expressed in the male germ line. Further characterization of the Piwi-family proteins in mammals will help to understand the biogenesis of tel-sRNAs. Profound strand asymmetry of some piRNAs and their lack of Dicer dependence prompt the view that piRNAs are not generated from dsRNA precursors. Instead, a ping-pong model involving single strand RNA transcripts was proposed for the generation of piRNAs in Drosophila, as well as prepachytene piRNAs in mice (Aravin et al. 2007a; Hartig et al. 2007; Klattenhoff and Theurkauf 2008). Heterogeneous single-strand noncoding RNA transcripts (TelRNA or TERRA) were reported to associate with telomeres in mice (Horard and Gilson 2008). These transcripts share the same strand specificity as tel-sRNA, consisting of UUAGGG repeats. While tel-sRNAs are not likely a result of either abortive transcription or the random degradation of lone telomere RNA transcripts, the biogenesis of tel-sRNAs warrants future investigation. In particular, it will be interesting to test whether long telomere RNA transcripts are the precursors to tel-sRNAs.
The change in the abundance of tel-sRNA in both Mll(−/−) and Suv39h1/h2(−/−) cells (Fig. 3A) suggests that tel-sRNAs are likely regulated by epigenetic mechanisms. It is well established in fission yeast that heterochromatin-specific histone modifications promote small RNA production from heterochromatin through a self-reinforcing loop. Our results showing the positive correlation between the levels of tel-sRNAs and the heterochromatin status at telomeres are indicative of a similar interaction between small RNAs and chromatin marks at telomeres. Recently, it was reported that an 18-mer RNA oligo of (UUAGGG)3 can inhibit telomerase TERT activity in vitro (Schoeftner and Blasco 2008), possibly by RNA duplex formation in the template region of the telomerase RNA component (TERC). If this is true in vivo, then tel-sRNAs, which contain UUAGGG repeats, could potentially act as sensors of chromatin status and complete a feedback loop between the telomere length control and telomeric heterochromatin regulation. This is consistent with the observations that Suv39h1/h2(−/−) cells, which have aberrantly long telomeres, have drastically lower level of tel-sRNAs (Fig. 3A) and higher TERT activity (Garcia-Cao et al. 2004). Future studies on the biogenesis and functions of tel-sRNAs will be key, to understand the role played by tel-sRNAs in the telomere length control.
MATERIALS AND METHODS
Cell lines, strains, and culture conditions
Mll(+/+) and Mll(−/−) MEF cells were isolated from 13.5-d-post-coitum embryos, immortalized MEF cells were generated by low oxygen incubation. Wild-type ES cells AB 2.2 (Naya et al. 1997) and Dicer(−/−) ES cells were grown on plates with either a layer of STO feeder cells or gelatin coating.
Wild-type CU427, CU428 stains and Dcl1(−/−) strains of Tetrahymena were grown in SPP medium at 30°C. For conjugation, log-phase cells of different mating types were starved (16–24 h) and mixed for various time.
Small RNA Northern blot and hybridization probes
Total RNAs were isolated using TRIzol reagent (Invitrogen) and small RNAs were further enriched using the Purelink miRNA isolation kit (Invitrogen). DNase I (Ambion) digestion was performed at 37°C for 25 min. For Northern blots, 2–3 μg RNAs were separated on a 12% or 15% polyacrylamid/urea/1XTBE gel and transferred to a Hybond-N positively charged membrane (GE) at 2 mA/cm2 for 75 min. Northern hybridization was carried out in Church buffer (0.25 M sodium phosphate buffer at pH 7.2, 1mM EDTA at pH 8.0, 1% BSA, 7% SDS) at 42°C for 16 h followed by three washes in 2X SSC 0.1% SDS at 42°C for 5 min and once in 1X SSC 0.1% SDS at 42°C for 10 min.
Hybridization probes for Tetrahymena small RNA blot are: G probe, 5′-(TGGGGT)3-3′; C probe 5′-(ACCCCA)3-3′; M element probe, 5′-TGTTAGAATTTATCAGATCTACCTGTGATTTTATTAAAAAGAAAGGCCTT-3′. For ES cells, probes are 5′-ACCCTAACCCTAACCCTA-3′; tRNA-Ile-AAT, 5′-CTTGGCGTTATTAGCACCAC-3′; and miR-293, 5′-ACACTACAAACTCTGCGGCACT-3′, respectively.
All probes were end labeled with γ-32P ATP by T4 polynucleotide kinase (NEB). An end-labeled RNA decade marker (Ambion) was used as the size marker.
Perioxidation/β-elimination reaction
For β-elimination, DNase I treated RNAs were dissolved in 17.5 μL borax buffer at pH 8.6 (4.375 mM borax [Fluka], 50 mM boric acid [Sigma]), and 2.5 μL 0.2 M sodium periodate (Sigma) was added. The reaction mixture was incubated for 10 min at room temperature in the dark and, after the addition of 2 μL glycerol it was incubated for an additional 10 min. The mixture was lyophilized using SpeedVac (Labconco), dissolved in 50 μL borax buffer at pH 9.5 (33.75 mM borax, 50 mM boric acid; pH adjusted by NaOH), and incubated for 90 min at 45°C. The samples were desalted and subject to Northern blot analysis. As a control, a 5′-end 32P-labeled RNA oligo was treated exactly the same way and separated on 12% gel. The sequence for the synthetic RNA oligo is 5′-AAGUGAUCUAAAGACCCAAGUUAUUAGUU-3′.
Quantitative RT-PCR for telomeric RNA
For small RNA RT-PCR analysis, total RNAs were treated with DNase I (Ambion) and subject to reverse transcription using a miScript reverse transcription kit (Qiagen). Specifically, the reverse transcriptase mix provided in the Qiagen kit includes a poly(A) polymerase. It adds poly(A) tail to small RNAs, which are not polyadenylated in vivo. The subsequent reverse transcription is carried out using an oligo-dT primer that have a universal tag sequence on the 5′ end. This universal tag allows amplification in the real-time PCR step. Using this approach, quantitative RT-PCR for tel-sRNAs was performed using telomere specific primer (TAGGGT)3 and miScript primer (anneal to the universal tag) provided by miScript SYBR Green PCR kit (Qiagen). All results were normalized against 5S RNA, which was detected by RT-PCR using random primers. The sequences for 5s rRNA primers are (1) forward: 5′-TCTACGGCCATACCACCCTGA-3′ and (2) reverse: 5′-GCCTACAGCACCCGGTATTCC-3′.
Cloning and sequencing tel-sRNAs
The PCR products from the RT-PCR analysis described above were purified by GIAquick Gel extraction kit (QIAGEN) and cloned into a TA cloning vector pCR2.1-TOPO (TOPO TA cloing, Invitrogen). Twenty clones were minimally prepared and subsequently sequenced using M13 reverse primer (5′-CAGGAAACAGCTATGAC-3′) in the TA vector. For analyzing the sequencing results, primer specific sequence and polyA sequence were identified. The sequence in between is indicative of the recovered tel-sRNA sequence. Given the repetitive nature of the telomere sequence, it is possible to have variations (no more than 6 nt) in the length of PCR products.
Stable knockdown of TRF1
Lenti-viruses expressing control and TRF1 shRNAs were obtained by transfecting 293FT cells with the pLKO.1-puro shRNA plasmid (Sigma-Aldrich, MISSION TRC shRNA NM_009352) or pLKO vector in addition to the pVSV-G and pCMVdelta8.2 helper plasmids. Virus-containing supernatant was harvested, and two sequential viral infections were performed on immortalized MEF cells (PD28) in the presence of 8 μg/mL hexadimethrine bromide. The MEF cells were then selected with 4 μg/mL puromycin and puromycin-resistant cells were pooled after 2 wk.
ChIP assay and telomere slot blots
Chromatin immunoprecipitations were performed following manufacturer's instructions (Millipore). Antibodies used were: anti-MLLc (Dou et al. 2005), anti-H3 trimethyl K4 (Abcam ab8580), anti-H3 trimethyl K9 (Millipore 17-625), anti-TRF1 (Abcam ab1423), and anti-RNA Pol II (Covance, MMS-129R). For slot blot, DNAs (10% inputs and immunoprecipitated DNAs) were blotted to a Hybond-N positively charged membrane (GE) and hybridized with α-32P labeled 1.6–2 kb PCR products generated by telomere oligos (TTAGGG)4 and (CCCTAA)4. Hybridization signals were quantified using ImageJ software. The relative intensity of immunoprecipitated DNA vs. input was calculated after extracting background signals, which is the no antibody control lane, from both samples.
For real-time PCR, ChIP DNA is reverse-crosslinked and subjected to quantitative PCR using primers specific to chromosome 1 and chromosome 11 as following: chromosome 1, 5′-TTAGGACTTCTGGCTTCGGTAG-3′ and 5′-AGCTGTGGCAGGCATCGTGGC-3′; chromosome 11, 5′-CACCCAGGCACAGATAGA-3′ and 5′-AGAAATGACCAGGCTCCA-3′. The PCR products were normalized to 5% input as previously described.
ACKNOWLEDGMENTS
We are grateful to Drs. Thomas Jenuwein and Gregory Hannon for providing us with the Suv39h1/h2(−/−) and Dicer(−/−) cells, respectively, and to Dr. Ming Lei for providing us with the cDNAs for TRF1, TRF2, and Tin2. We also thank Dr. Jay L. Hess for helpful discussions. This work was supported by funds from NIH (R01GM082856-01) and Nathan Shock Aging Center, University of Michigan, to Y.D.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1423309.
REFERENCES
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet. 2007a;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol. 2007b;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Kavi HH. Molecular biology. Slicing and dicing for small RNAs. Science. 2008;320:1023–1024. doi: 10.1126/science.1159018. [DOI] [PubMed] [Google Scholar]
- Blasco MA. Telomere length, stem cells, and aging. Nat Chem Biol. 2007a;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007b;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes & Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hartig JV, Tomari Y, Forstemann K. piRNAs—The ancient hunters of genome invaders. Genes & Dev. 2007;21:1707–1713. doi: 10.1101/gad.1567007. [DOI] [PubMed] [Google Scholar]
- Horard B, Gilson E. Telomeric RNA enters the game. Nat Cell Biol. 2008;10:113–115. doi: 10.1038/ncb0208-113. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes & Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJ. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Moazed D, Buhler M, Buker SM, Colmenares SU, Gerace EL, Gerber SA, Hong EJ, Motamedi MR, Verdel A, Villen J, et al. Studies on the mechanism of RNAi-dependent heterochromatin assembly. Cold Spring Harb Symp Quant Biol. 2006;71:461–471. doi: 10.1101/sqb.2006.71.044. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes & Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Huang H-P, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai M-J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes & Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- Stadler BM, Ruohola-Baker H. Small RNAs: Keeping stem cells in line. Cell. 2008;132:563–566. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol. 2006;71:487–496. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]