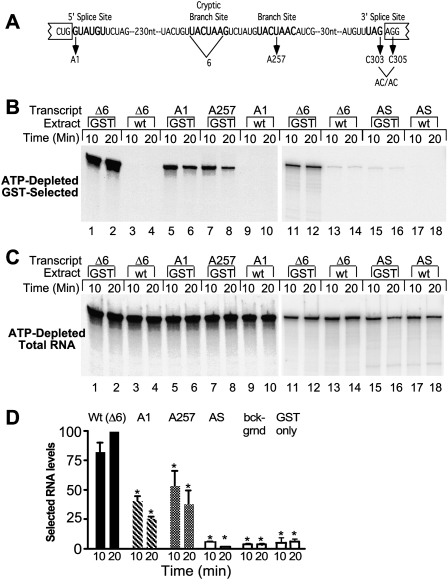
FIGURE 4.
Efficient association of Prp5p with RNA requires a functional intron. (A) Diagram of actin wild-type and mutant pre-mRNAs. (From left to right) Bold lettering indicates the 5′ splice site, the cryptic branch site, the authentic branch site, and the 3′ splice site. Also depicted are mutant pre-mRNAs and the 6-nt cryptic branchpoint sequence deleted from the Δ6 transcript. The A1, A257, and AC/AC mutations were each made in the Δ6 background. (B) RNA in GST-Prp5p selected material. ATP-depleted GST-Prp5p (GST) or wild-type (WT) extracts in splicing buffer were incubated with equal amounts of functional actin (Δ6), 5′ splice site mutant (A1), branch site mutant (A257), and antisense (AS) RNAs for the indicated times. Splicing reactions were subjected to GST selection after which RNA was extracted, fractioned by denaturing PAGE, and visualized by autoradiography (12-h film exposure for lanes 1–10 and 96-h PhosphorImager screen exposure for lanes 11–18). (C) Total RNA from splicing reactions. RNA was extracted from samples removed from reactions before GST selection. RNA was visualized by autoradiography (2.5-h film exposure for lanes 1–12 and 67-h Phos-phorImager screen exposure for lanes 11–18). (D) Quantification of GST-selected RNA levels. Levels of selected wild-type and mutant A1 and A257 RNAs in three experiments as in A were measured and calculated as percentage of wild-type RNA pulled down at 20 min in the same experiment. The means (±SD) are plotted. Levels statistically significantly different from Δ6 levels are indicated by asterisks; all p ≤ 0.01. The levels of A1 and A257 are not statistically different from each other. Antisense RNA levels are the same as background levels (selections with an untagged Prp5 protein and selections with GST-tag only) measured in two, 10, and two experiments, respectively.