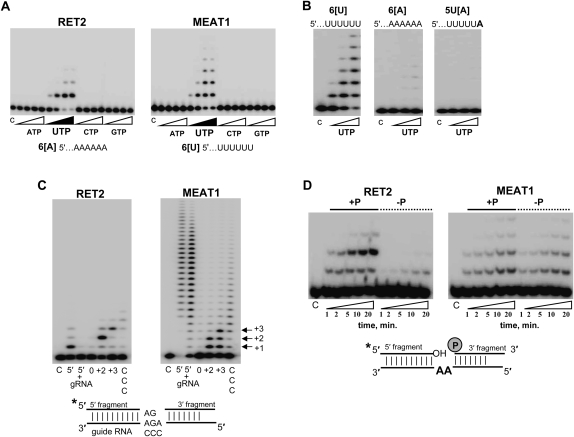
FIGURE 3.
Substrate specificity of the purified MEAT1. (A) Nucleotide triphosphate substrate specificity. C indicates control RNA. Reactions were carried out for 30 min in the presence of 1, 10, 100, and 500 μM of respective NTPs; 10 nM of each enzyme; and 100 nM of chemically synthesized RNAs. Products were separated on 15% acrylamide/8 M urea gels. (B) Effect of the terminal ssRNA nucleotides on MEAT1 polymerization activity. Reactions were set up as in A at 1, 10, and 100 μM of UTP. (C) Precleaved U-insertion editing assay. RNA substrate was assembled by heating and slow cooling of the radiolabeled 5′ fragment, guide RNA, and 3′ fragment and at 1:1.5:2 molar ratios, respectively. Guide RNAs were designed to direct zero, two, or three U-insertions or to have three cytidines in guiding positions. Reactions containing 100 μM of UTP, 100 nM of 5′ fragment, and 50 nM of enzyme were incubated for 30 min. 5′ indicates 5′ fragment; 5′+g, only 5′ fragment and gRNA were assembled; 0, +2, and +3, number of guiding nucleotides; and CCC, cytidines instead of guiding purines. (D) Reactions were done as in C for +2 guiding, except with the dephosphorylated 3′ fragment in the “–P” panels.