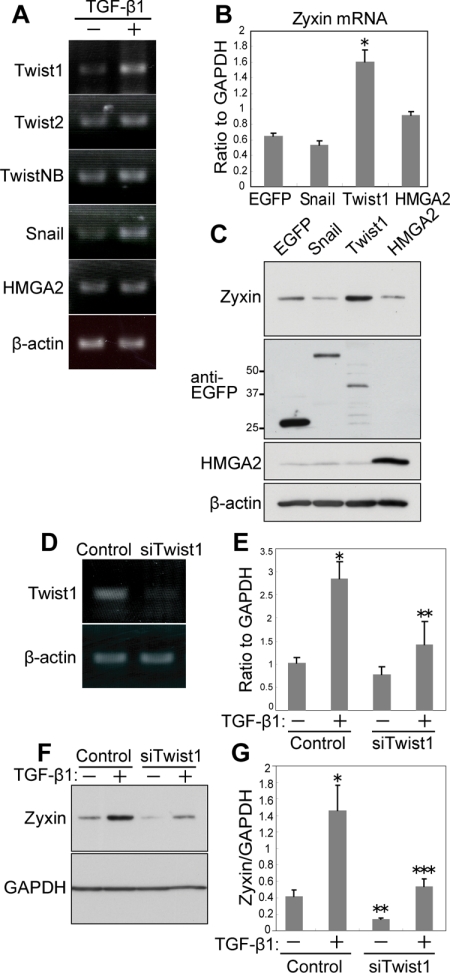
Figure 5.
Twist1 regulates zyxin in TGF-β1-EMT. (A) Expression analysis of EMT inducers in NMuMG-C7 cells. RT-PCR performed with total RNA isolated from NMuMG-C7 cells treated with or without TGF-β1. β-Actin was used as a control. (B) Twist1 up-regulates zyxin. Real-time PCR performed with total RNA from NMuMG-C7 cells transduced with EGFP, Snail, Twist1, or HMGA2. *p = 0.0122 versus EGFP. Error bars, the SEM of three technical replicates. (C) Twist1 induced the expression of zyxin. Immunoblot analysis of zyxin in NMuMG-C7 cells transduced with EGFP, EGFP-Snail, EGFP-Twist1, or HMGA2. The expression of transduced genes was confirmed by blotting with anti-EGFP and anti-HMGA2 antibodies. (D) Efficacy of Twist1 RNAi assessed by RT-PCR. Total RNA from NMuMG-C7 cells transfected with either Twist1 siRNA or scramble RNA were subjected to RT-PCR. (E) Twist1 depletion abrogated up-regulation of zyxin by TGF-β1. Real-time PCR analysis of zyxin mRNA in NMuMG-C7 cells transfected with Twist1 siRNA or scramble RNA and treated or not with TGF-β1. Results are shown as the mean ± SEM (n = 3). *p = 0.011 when compared with control, TGF-β1(−); **p = 0.0236 when compared with control, TGF-β1(+). (F) Twist1 depletion suppressed the expression of zyxin. Immunoblot analysis of zyxin in NMuMG-C7 cells transfected with Twist1 siRNA or scramble RNA and treated or not with TGF-β1. GAPDH was used as a loading control. (G) Results of F were quantitated by densitometry using NIH ImageJ. Error bars, SD from triplicate experiments. *p = 0.0084 when compared with control, TGF-β1(−); **p = 0.035 when compared with control, TGF-β1(−); ***p = 0.012 when compared with control, TGF-β1(+).