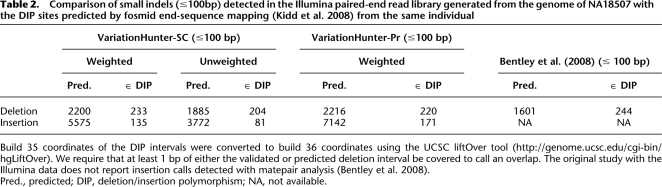
Table 2.
Comparison of small indels (≤100bp) detected in the Illumina paired-end read library generated from the genome of NA18507 with the DIP sites predicted by fosmid end-sequence mapping (Kidd et al. 2008) from the same individual
Build 35 coordinates of the DIP intervals were converted to build 36 coordinates using the UCSC liftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver). We require that at least 1 bp of either the validated or predicted deletion interval be covered to call an overlap. The original study with the Illumina data does not report insertion calls detected with matepair analysis (Bentley et al. 2008).
Pred., predicted; DIP, deletion/insertion polymorphism; NA, not available.