Abstract
The Apicomplexa are a group of phylogenetically related parasitic protists that include Plasmodium, Cryptosporidium, and Toxoplasma. Together they are a major global burden on human health and economics. To meet this challenge, several international consortia have generated vast amounts of sequence data for many of these parasites. Here, we exploit these data to perform a systematic analysis of protein family and domain incidence across the phylum. A total of 87,736 protein sequences were collected from 15 apicomplexan species. These were compared with three protein databases, including the partial genome database, PartiGeneDB, which increases the breadth of taxonomic coverage. From these searches we constructed taxonomic profiles that reveal the extent of apicomplexan sequence diversity. Sequences without a significant match outside the phylum were denoted as apicomplexan specialized. These were collated into 9134 discrete protein families and placed in the context of the apicomplexan phylogeny, identifying the putative origin of each family. Most apicomplexan families were associated with an individual genus or species. Interestingly, many genera-specific innovations were associated with specialized host cell invasion and/or parasite survival processes. Contrastingly, those families reflecting more ancestral relationships were enriched in generalized housekeeping functions such as translation and transcription, which have diverged within the apicomplexan lineage. Protein domain searches revealed 192 domains not previously reported in apicomplexans together with a number of novel domain combinations. We highlight domains that may be important to parasite survival.
The Apicomplexa is a protozoan phylum of around 5000 species, the majority of which are parasitic, infecting a wide range of animals from mollusks to mammals (Cavalier-Smith 1993). Many species of Apicomplexa cause diseases of medical and veterinary importance and represent a significant economic burden and global healthcare challenge. The emergence of parasite strains resistant to the few efficacious treatments that are available underscores the urgent need to identify new classes of drug targets and novel antiparasitic therapeutics (Aspinall et al. 2002; Trouiller et al. 2002; White 2004). Members of the phylum include: Plasmodium, the etiological agent of malaria, accounting for one-in-five deaths among children under the age of five in Africa (World Health Organization 2003); Toxoplasma gondii, the causative agent of toxoplasmosis, infects nearly one-in-three of the adult population with severe implications for those living with HIV/AIDS (Belanger et al. 1999); Cryptosporidium, a waterborne pathogen also with implications for immune-compromised individuals (Hunter and Nichols 2002); the invertebrate parasite Gregarina, a useful model for studying apicomplexan motility; and the agricultural parasites Eimeria, Neospora, Sarcocystis, and Theileria, which cause disease across a range of livestock (Graat et al. 1996; Dubey 1999).
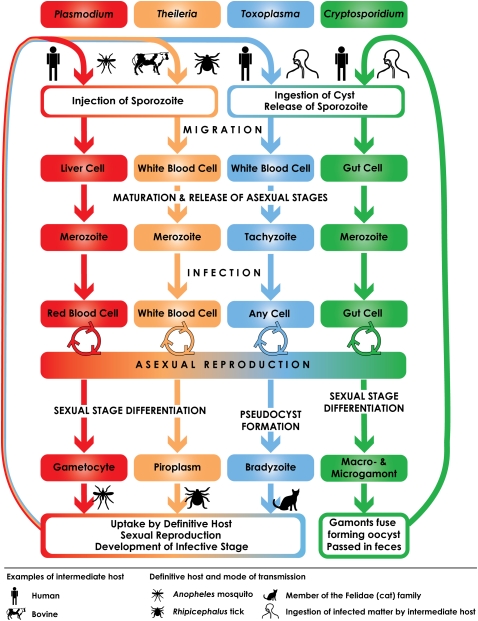
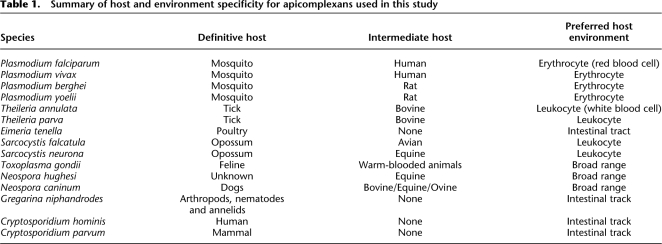
The apicomplexan life cycle is complex and may be broken down into three broad stages: sporozoite, merozoite, and gametocyte (Fig. 1). While the general life cycle is common to the phylum, there are striking differences between species. Some require a single host (e.g., Cryptosporidium), whereas others are more complex, requiring sexual reproduction in the vector species for transmission (e.g., Theileria and Plasmodium; Table 1). Apicomplexans are characterized by a number of defining organelles involved in host cell attachment, invasion, and the establishment of an intracellular parasitophorous vacuole within the host cell. The arsenal of organelles varies between species, but typically includes rhoptries, micronemes, and dense granules. Proteins stored in these vesicles are released through the apical complex at the anterior of the cell (Sibley 2004). In addition, all apicomplexans examined to date (with the exception of the Cryptosporidium and Gregarina) possess an apicoplast organelle that is hypothesized to be an ancient secondary endosymbiosis with an algal cell (Zhu et al. 2000b; Fast et al. 2001; Toso and Omoto 2007). The genome of this plastid has reduced to the point where the remaining genes are predominantly involved in organelle replication (Wilson et al. 1996). Nonetheless, many additional proteins involved in three critical metabolic pathways: fatty acid biosynthesis, isoprenoid biosynthesi, and for Plasmodium, haem degradation (Ralph et al. 2004; Waller and McFadden 2005) are targeted to the apicoplast. Due to their essential role, many enzymes involved in these pathways are the focus of drug target discovery programs.
Figure 1.
Apicomplexan life cycles. Members of the Apicomplexa share a generalized life cycle, though each species has its own specializations. Plasmodium spp. and Theileria spp. are transmitted and undergo sexual recombination in an insect vector, the Anopheles mosquito and Rhipicephalus tick, respectively. Cryptosporidium is able to autoinfect its host; the oocyst can sporulate and excyst in the same host, maintaining the infection for months to years. The Coccidian parasites are represented in this figure by Toxoplasma, which is able to infect the majority of warm-blooded animals. The differentiation of Toxoplasma tachyzoites into gametocytes is triggered only when members of the cat family (Felidae) are infected. The molecular basis for this regulation is not yet known. The intermediate and definitive host spectrum for each species under consideration in this study are given in Table 1.
Table 1.
Summary of host and environment specificity for apicomplexans used in this study
To develop novel antiparasitic compounds and increase our understanding of apicomplexan biology, several large-scale-sequencing projects have been initiated. The complete genomes of six apicomplexans are currently available (Fig. 2; Carlton et al. 2002; Gardner et al. 2002, 2005; Abrahamsen et al. 2004; Xu et al. 2004; Pain et al. 2005). Furthermore, large expressed sequence tag (EST) collections have been generated for an additional nine species (Ajioka et al. 1998; Howe 2001; Li et al. 2003a; Cui et al. 2005), which can be used to construct so-called “partial genomes” (Peregrin-Alvarez and Parkinson 2007; Wasmuth et al. 2008). Studies of these data sets have provided insights into adaptations that relate to their parasitic ecology, such as the identification of expanded families of proteases in Plasmodium (Wu et al. 2003). More systematic sequence comparisons have started to uncover genes restricted to specific Apicomplexa orders (Li et al. 2003a; Martens et al. 2008), or even individual species (Brayton et al. 2007).
Figure 2.
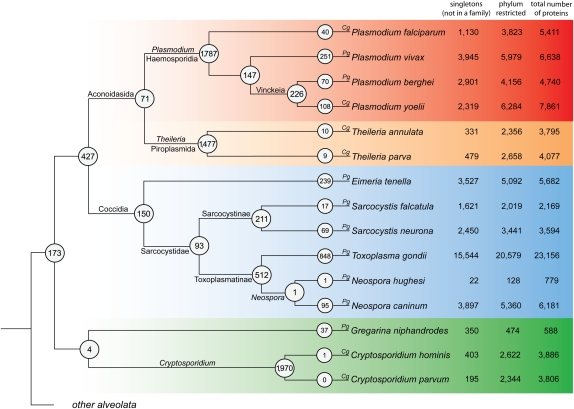
Apicomplexan-specialized protein families. This figure shows the putative point of origin for each protein family within the Apicomplexa. The species membership for each protein family was used to determine the putative point of origin of protein families within the apicomplexan phylogeny. The number of protein families shared between at least two daughter taxa of a particular clade is circled at each node. On the terminal branches is the number of species-specific protein families, where a family contains at least two proteins. The number of singletons (not clustered in a family), total number of phylum-restricted proteins, and total number of proteins available are given after the species name. Whether a complete or partial genome is available for the species is designated with a Cg or Pg, respectively. The tree is a cladogram, and the construction of the phylogeny is described in the Methods section.
The availability of genomic data sets for 15 species offers an opportunity to build on these initial studies and undertake more systematic analyses to explore genetic diversity within the Apicomplexa. Here, we catalog the taxonomic profile of each apicomplexan species and identify those that share little or no similarity to nonapicomplexan proteins. From this set of “apicomplexan-specialized” sequences, protein families are constructed and placed within the context of the apicomplexan phylogeny. This highlights the evolutionary history of proteins associated with a parasitic lifestyle and identifies groups of proteins with functional annotations that shed light on apicomplexan biology. To authenticate proteins that are otherwise annotated as “hypothetical,” we compare our results with the proteomic data sets that are available for five of the species. Finally, domain analyses identified both the taxonomic distribution of apicomplexan domains as well as domain architectures specific to the Apicomplexa.
Results
Taxonomic distribution of apicomplexan proteins
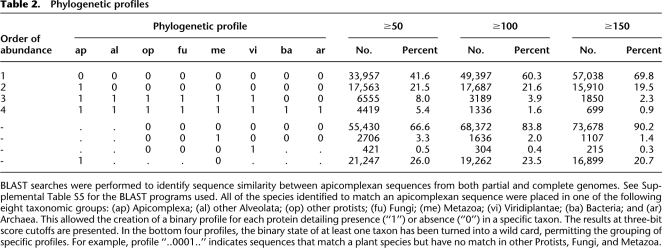
To identify the levels of sequence conservation between the Apicomplexa and other taxa, a series of BLAST searches were performed against the following databases (selected to maximize sequence and taxonomic coverage): COGENT for complete genomes, UniProt for nonredundant proteins, and PartiGeneDB for partial genomes. In total, 87,736 apicomplexan sequences were compared with ∼6.6 million sequences from 1112 species (including 501 eukaryotes and 552 bacteria). For each apicomplexan sequence, the taxonomic source of the database hits were assigned to one of eight divisions: (1) other Apicomplexa, (2) other Alveolata, (3) other Protista, (4) Fungi, (5) Metazoa, (6) Viridiplantae, (7) Bacteria, and (8) Archaea. By combining the scores from each of the divisions, “taxonomic profiles” were created using a binary classification scheme. For example, the profile “00110010” indicates an apicomplexan sequence with homology with sequences from other Protista, Fungi, and Bacteria. To provide a comprehensive view of the relationships between apicomplexan sequences and other taxa, we used three BLAST bit score cutoffs to generate taxonomic profiles. For a discussion on the use and impact of BLAST score cutoffs, see Supplemental File S1.
The four most common profiles were associated with the extremes of phylogenetic diversity (Table 2). Sequences specific to a single species or matching only other apicomplexans (profiles: 00000000 and 10000000, respectively) were the most abundant, accounting for 63%–89% of all sequences, depending upon bit score cutoff. The next two most abundant profiles represent the other end of the diversity spectrum, being either highly conserved across all eukaryotes (profile: 1111100) or the three domains of life (profile: 11111111). Discussion on the functions of these proteins is presented in Supplemental File S1.
Table 2.
Phylogenetic profiles
BLAST searches were performed to identify sequence similarity between apicomplexan sequences from both partial and complete genomes. See Supplemental Table S5 for the BLAST programs used. All of the species identified to match an apicomplexan sequence were placed in one of the following eight taxonomic groups: (ap) Apicomplexa; (al) other Alveolata; (op) other protists; (fu) Fungi; (me) Metazoa; (vi) Viridiplantae; (ba) Bacteria; and (ar) Archaea. This allowed the creation of a binary profile for each protein detailing presence (“1”) or absence (“0”) in a specific taxon. The results at three-bit score cutoffs are presented. In the bottom four profiles, the binary state of at least one taxon has been turned into a wild card, permitting the grouping of specific profiles. For example, profile “..0001..” indicates sequences that match a plant species but have no match in other Protists, Fungi, and Metazoa.
Introducing a wild-card (‘.’) character for searching the profiles adds flexibility. Depending on the BLAST cutoff, we identified from 215 to 421 sequences with matches to Viridiplantae, to the exclusion of other eukaryote phyla (profile: 0.00001..). These sequences may represent possible homologs of algae origin, translocated from the early apicoplast genome (Huang et al. 2004). A subset of these sequences was associated with a range of metabolic functions. Of these, we noted that restricted to the Theileria were eight choline kinase proteins: an enzyme involved in lipid metabolism and implicated in host cell transformation (Gardner et al. 2005). A phylogenetic reconstruction of the parasite proteins with other choline kinases from UniProt placed many of the Theileria proteins into a single clade (data not shown). How these parasite enzymes affect the host cell is not clear; these proteins do not contain canonical signal peptides, while only one possessed a single predicted transmembrane region.
Apicomplexan-specialized protein families
Given the large proportion of sequences lacking putative homologs outside of the Apicomplexa, we were interested in identifying how this novelty was distributed throughout the phylum. To address this, we used TribeMCL (Enright et al. 2002) to group sequences into families. A score cutoff of 100 bits was used to collate apicomplexan-specific sequences. Here, the term apicomplexan-specialized denotes proteins unique to the taxon as well as proteins that have undergone such a large amount of divergence as to fall below our bit score cutoff. Of 67,015 apicomplexan-specialized proteins, 27,901 (41%) were assigned into one of 9134 families, where a family contains at least two members. We refer to proteins not assigned a family as singletons. Each family was then mapped in the context of known phylogenetic relationships to identify putative points of origin and examine species and taxon-specific protein family expansions (Fig. 2).
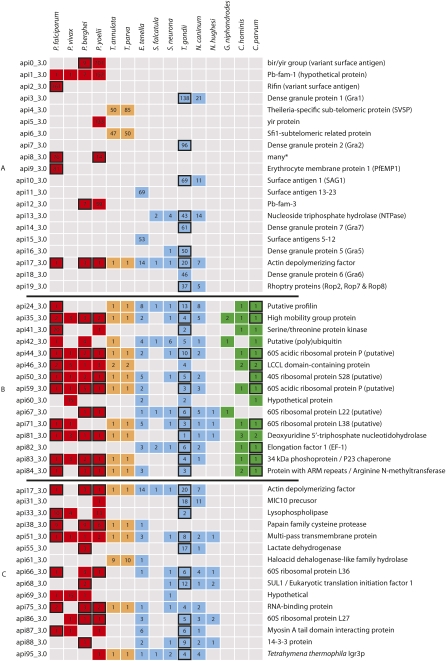
Most protein families are restricted to a single genus, highlighting the huge proteomic diversity underlying their disparate biology. Indeed, 19 of the 20 most abundant families were present in a small number of closely related taxa, or were species specific (Fig. 3A). Typically, these families were well-known apicomplexan specializations, many of which have been shown to be necessary for parasite survival in the host. However, we also identified ancestral families common to wider groups of Apicomplexa. In the following, we concentrate on protein families associated with infection machinery, focusing on three clades: Aconoidasida, Coccidia, and pan-Apicomplexan. Analyses performed on additional families are available in Supplemental Text File S1.
Figure 3.
Species distribution of sequences in a selection of Apicomplexan-specialized protein families. Three sets of protein families are presented: (A) the top 20 most-abundant families; (B) the top 15 most-abundant families with sequence representatives from the 173 families conserved between the Cryptosporidium and either or both of the Aconoidasida and the Coccidia; and (C) the top 15 most-abundant families, with sequence representatives from the 427 families conserved between the Aconoidasida and the Coccidia. Numbers in boxes indicate the number of sequences from each species associated with each family. A black border for a box shows that proteomic data can be mapped to the protein family. The colors refer to the clades in Figure 2.
Protein families within the Aconoidasida
The life cycles of the Haemosporidia and Piroplasmida are broadly similar, both occupying red blood cells, which assist their transmission to their respective insect vectors (Fig. 1; Table 1). However, given the large differences in the rest of the life cycle, including invasion of leukocytes by Theileria, strategies for immune evasion, choice of vector species, and host cell modification, it is perhaps not surprising that only 71 of the 4196 families associated with this group were conserved across the Aconoidasida (Supplemental Table S1).
Interestingly, six of the 20 largest protein families within the Apicomplexa are examples of variant Plasmodial antigens: BIR/VIR (api0_3.0 and api5_3.0), RIFIN (api2_3.0), and PfEMP1 (api9_3.0). These antigens are presented on the surface of infected red blood cells and are thought to interfere with the host immune response. The other large Plasmodium families are the Pb-fam groups (api1_3.0 and api12_3.0), which were first annotated in the P. berghei genome (Hall et al. 2005); we show that these are present throughout the genus and have possibly undergone a large expansion in P. yoelii.
Protein families within the Coccidia
There were 2236 protein families restricted to the Coccidia. Of these, 512 were shared between Neospora and Toxoplasma, consistent with their close evolutionary relationship and similar ecology (Figs. 1, 2). Of the 150 families shared across Eimeria and the Sarcocystidae, 25 were found in at least three species (Supplemental Table S2).
The micronemes and rhoptries are the first organelles to secrete their protein complements, starting the invasion process. These proteins are implicated in attachment to the host cell, leading to the formation of the moving junction, through which the parasite enters the host cell. Microneme proteins include those involved with host cell adhesion (e.g., MIC1 and MIC2) and a class of four aspartyl proteases termed Toxomepsins. Although the precise functions of Toxomepsins have yet to be refined, work in Eimeria has shown Toxomepsin 3 to be localized to the parasite's apical tip shortly after cell attachment (Jean et al. 2000), with signal peptides present in Toxomepsins 2 and 3 (Dowse and Soldati 2004). While Toxomepsin 3 was present across the Coccidia, Toxomepsins 1 and 2 were specific to the Toxoplasmatinae.
With the moving junction formed, the rhoptries discharge their contents. The largest group of rhoptry proteins is the ROP2 family, which are restricted to Toxoplasma and N. caninum (api19_3.0). The ROP2 family contains the closely related ROP2, ROP4, ROP7, and ROP8 proteins (El Hajj et al. 2006), which, in our analysis, were clustered into two inclusive families. Fewer rhoptry proteins could be annotated in the other coccidian data sets. In Sarcocystis, we found only ROP17 and ROP18. Like the rhoptry neck proteins found in Eimeria, (RON2 and RON4), these represented species-specific families. The high level of sequence divergence of rhoptry proteins may explain the putative absence of other rhoptry protein homologs in Sarcocystis and Eimeria. However, there are two further possible explanations. Firstly, Sarcocystis and Eimeria ROP homologs may be expressed at lower levels than their Toxoplasmatinae counterparts, reducing our ability to detect them within the EST data sets. Secondly, in contrast to micronemes, where some proteins are conserved throughout the Apicomplexa (Kappe et al. 1999), our analysis showed that of the 192 families annotated as rhoptry protein families, only three were present in more than one genus (or Toxoplasma and Neospora). We also noticed distinct families for both Theileria and Cryptosporidium, and so it is probable that Sarcocystis and Eimeria have their own additional battery of rhoptry proteins that await discovery.
Among the largest families in Coccidia were those associated with the dense granule (DG) organelles. These include GRA proteins that were found to share little sequence similarity across the families. Due to its amenability to genetic investigation, most studies of GRA proteins have focused on their role in Toxoplasma, where they are associated with either the membrane of the PV or the membranous nanotubular network, which connects the parasite with the PV membrane. Homologs to some Toxoplasma GRA proteins have been identified in N. caninum (Mercier et al. 2005). We were able to identify GRA1–GRA8 and GRA10 in Toxoplasma, and orthologs for GRA1, GRA2, GRA4, and GRA8 in Neospora. To the best of our knowledge, Neospora GRA4 and GRA8 proteins have not been previously reported. The absence of GRA proteins in the Eimeria and Sarcocystis data sets supports the possibility that dense granules are either absent or function differently in these organisms (Daszak et al. 1993). These findings suggest a relatively recent development of GRA proteins in the Toxoplasmatinae.
Pan-apicomplexan protein families
From Figure 2, the vast majority of protein families specific to the Apicom-plexa represent species- or genus-specific innovations. This supports the notion that the Apicomplexa as a group do not share many ancient common adaptations (Martens et al. 2008). Six hundred of the 9134 families were shared across deeper taxonomic groupings, of which 173 protein families were identified as potentially being present in the ancestral apicomplexan (Fig. 3B; Supplemental Table S3). The largest protein family (api24_3.0) contained 36 proteins from 10 species and was annotated as containing a profilin domain, a component of the regulatory mechanism for parasite gliding motility. There is a high level of conservation within the apicomplexan profilins, but they are divergent from those in other eukaryotes (Baum et al. 2006). Profilin acts as a ligand for the murine Toll-like receptor TLR11, causing levels of IL-12 cytokine to be up-regulated, thereby eliciting an inflammatory response (Yarovinsky et al. 2005). The presence of profilin across the Apicomplexa suggests that metazoans have evolved a receptor capable of raising an immune response to this broad-spectrum antigen.
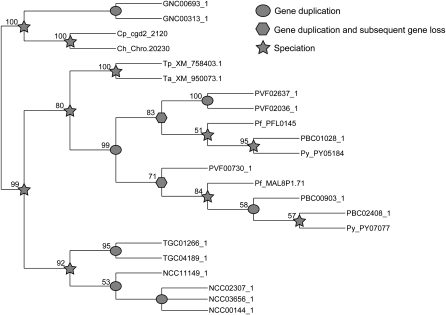
Ribosomal proteins were common in the largest families (Fig. 3B; Supplemental Table S3). While their classification as apicomplexan-specialized proteins may be surprising, inspection of their alignments expose variation between apicomplexan ribosomal proteins and those for other species (Supplemental File S1). Similar findings were observed for a group of chromatin-related factors annotated as “high mobility group” proteins (Briquet et al. 2006). Numerous gene duplication events have occurred in the Plasmodium and Toxoplasmatinae lineages, revealing potential differences in transcription regulation associated with each parasitic life cycle (Fig. 4).
Figure 4.
Proposed evolution of family 35_3.0. The multiple sequence alignment and phylogenetic reconstruction is described in the Methods section. There were two instances where gene duplication occurred, along with evidence for subsequent gene loss in one lineage. The incomplete nature of partial genomes makes interpretation of gene loss difficult, but has less effect when considering duplication events. Bootstrap values are given at the nodes and are a percentage of 1000 replicated multiple sequence alignments. Nodes supported by <50% of replicates are shown as a polytomy.
Apicoplast proteins
The apicoplast is an organelle acquired through the endosymbiosis of cyanobacteria, and is essential to the survival of the apicomplexan parasite, except for Cryptosporidium, which appears to have lost this plastid. A previous bioinformatic analysis of the P. falciparum genome predicted 545 proteins are imported into the apicoplast (Ralph et al. 2004). Many of these are involved in the three metabolic pathways: fatty acid biosynthesis, isoprenoid biosynthesis, and for Plasmodium, haem degradation. Of the 545 apicoplast proteins, 471 were found to possess homologs in other apicomplexans (bit score ≥ 100). The remaining 74 proteins specific to P. falciparum were also annotated as hypothetical. Considering the proteins with potential orthologs, 341 were restricted to the Plasmodium (P. falciparum plus another), including members of the haem biosynthetic pathway. No members of this pathway were found in other apicomplexans. The absence of enzymes involved in haem biosynthesis in nonplasmodial species was expected; Plasmodium is the only apicomplexan in this study whose asexual cycle degrades haemoglobin, providing an essential source of amino acids and iron. We manually checked a reported case of uroporphrinogen III synthase in Toxoplasma and consider it a misannotation (see Supplemental File S1).
In the Coccidia, we found enzymes involved in fatty acid synthesis, but, intriguingly, no members of the isopentenyl diphosphate biosynthesis pathway (Isp enzymes). This pathway is important in the modification of transfer RNAs, suppressing premature stop codons and frameshifts in coding regions (Petrullo et al. 1983). These coding errors have been found in Toxoplasma, Eimeria, and Neospora and Plasmodium (Lang-Unnasch and Aiello 1999; Cai et al. 2003). The absence of this pathway in the Coccidian data sets may reflect EST sampling biases. It is noted that the Isp enzymes are expressed at relatively low levels within P. falciparum (data not shown), which may affect their representation within the partial genome data sets.
Expanding the repertoire of protein domains to the Apicomplexa
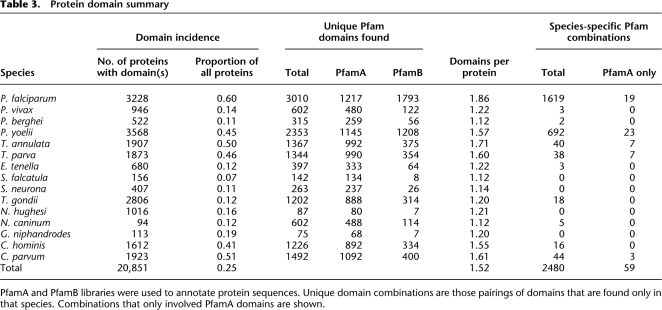
A more sensitive form of functional annotation is the decoration of sequence with protein domains. We used the Pfam database (Finn et al. 2006), considering both PfamA and PfamB divisions of the domain library. In total, 20,854 sequences were annotated with at least one protein domain (18,331 with PfamA and 4388 with PfamB; Table 3). Considering only PfamA domains, our searches revealed 192 known protein domains that had not been previously reported in the Apicomplexa. Forty-one (21%) of these domains were present in more than one species, many restricted to the Coccidia. The Kunitz trypsin inhibitor domain (PF00014) occurs in tandem repeats in N. caninum and T. gondii proteins. This domain is widespread in the Metazoa and found in a small number of bacteria and the Amsacta moorei entomopoxvirus. We believe this is the first report of this domain in a single-celled eukaryote, although its role in host immune evasion awaits further characterization.
Table 3.
Protein domain summary
PfamA and PfamB libraries were used to annotate protein sequences. Unique domain combinations are those pairings of domains that are found only in that species. Combinations that only involved PfamA domains are shown.
Among the Sarcocystidae were multiple instances of the annexin domain (PF00191). The domain has been implicated with a range of functions. In the diplomonad parasite Giardia lamblia, it is thought to regulate cytoskeletal dynamics during cyst transition (Bauer et al. 1999). From the coccidian EST libraries, annexin is only expressed in the infective tachyzoite phase and not the sessile bradyzoite form (Fig. 1). This suggests that one or more copies of the annexin domain found in these cyst-forming coccidians may have an analogous role to that of Giardia.
Present in the apicomplexan domain annotations was the Sushi domain (PF00084), also termed complement control protein module. These domains are associated with the regulation of complement activation, a critical component of the innate immune response. The domain probably arose early in the metazoan lineage and has subsequently been acquired by viruses (Ciulla et al. 2005). Our searches identified seven instances of the Sushi domain within the Apicomplexa: two in C. parvum (Templeton et al. 2004), C. hominis, and N. caninum, and one in T. gondii. A single Sushi domain was previously reported in P. falciparum, whose sequence can be aligned with the domain model (O'Keeffe et al. 2005). The apicomplexan Sushi domains were aligned with counterparts from the Metazoa and viruses. While the invariant four cysteines were present in the apicomplexan sequences, the parasite domains substitute the highly conserved tryptophan located near the C terminus, with a tyrosine or phenylalanine. Phylogenetic reconstruction of this domain has previously proven difficult (Soares et al. 2005), so to investigate relationships between the sequences, all-against-all pairwise alignments were generated, and the scores were hierarchically clustered. The parasite sequences were placed in a clade with domains from regulators of complement activity (RCA) proteins, including C4b-binding protein and complement receptor type 1 (Supplemental Fig. S2A). We found signal peptides in all full-length proteins containing the Sushi domain, as well as a C-terminal transmembrane domain in three proteins (Supplemental Fig. S2B), giving a potential role in the regulation of the host immune response.
Protein domain specializations within the Apicomplexa
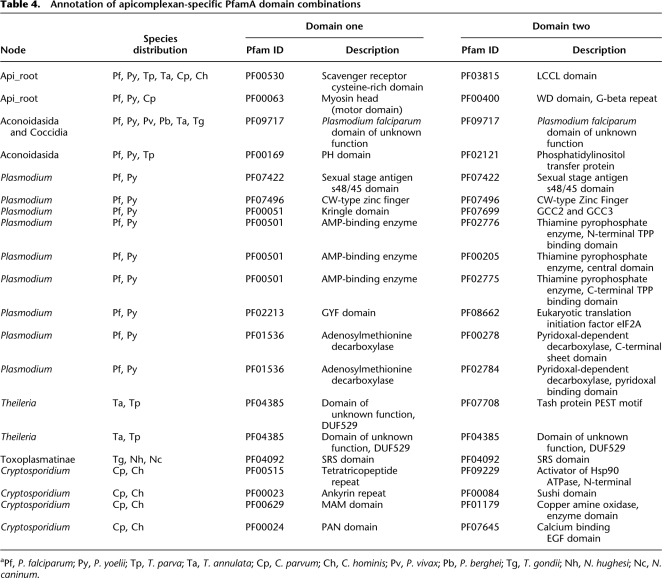
Within the PfamA database are 30 apicomplexan-specific domains. Our initial searches were only able to increase the species distribution of three of these domains. This is consistent with our earlier findings that the majority of apicomplexan-specific innovations appear to occur at the level of individual species and/or genera. We next examined the potential of novel domain couplings to increase parasite diversity. Most novel pairings involved PfamB domains. For example, considering both PfamA and PfamB annotations, 3247 domain pairings were unique to the Apicomplexa. Of these, 2480 (76%) were restricted to a single species (Table 3; Supplemental Fig. S3). Most of those domain partnerships found in two or more species were restricted to the Haemosporidia (Supplemental File S1).
Of domain combinations involving both PfamA and PfamB, 61 likely represent inventions in the ancestral Apicomplexa. We matched GO slim annotations (Ashburner et al. 2000) to these domains and, while the majority of domains could not be assigned a functional term, 12 terms were significantly enriched, including “pathogenesis,” “symbiosis,” and “interspecies interaction” (P < 0.001 for all three terms; Supplemental Table S5). Limiting the search to PfamA combinations, 20 pairs were found in at least two apicomplexans (Table 4; Supplemental Fig. S3) and two combinations could be considered pan-phylum. The PxSR protein (containing a scavenger receptor cysteine-rich domain [PF00530] and LCCL domain [PF03815]) has been implicated in oocyst differentiation into sporozoites in P. berghei (Claudianos et al. 2002). The second combination (Myosin head [PF00063] and WD repeat [PF00400]) has been shown to be located in the myonemes, structural filaments that form rings along the length of Gregarines (Heintzelman and Mateer 2008). However, the function of this protein in any apicomplexan has yet to be described.
Table 4.
Annotation of apicomplexan-specific PfamA domain combinations
aPf, P. falciparum; Py, P. yoelii; Tp, T. parva; Ta, T. annulata; Cp, C. parvum; Ch, C. hominis; Pv, P. vivax; Pb, P. berghei; Tg, T. gondii; Nh, N. hughesi; Nc, N. caninum.
Discussion
Here, we describe the most comprehensive analysis of protein family diversity across the Apicomplexa performed to date. This study builds on previous studies, for which fewer ESTs were available for fewer species or were restricted to complete genomes. For example, Li and coworkers performed a phylogenetic study of sequences from five apicomplexans and suggested that ∼2% of assemblies would form species-specific protein families (Li et al. 2003a). With many more sequences available for these and additional species, we can update this proportion to 7% of proteins. A more recent study, which included the six fully sequenced apicomplexan genomes, focused more on differences between the basal chromalveolate groups (Oomycetes, Diatoms, Apicomplexa, and Ciliates) (Martens et al. 2008). We concentrate specifically on the Apicomplexa and focus on extracting the biological implications of our findings. Use of the partial genomes expanded sequence space within the Plasmodium, and for the first time has allowed a detailed look at those protein families that are found only in the Coccidia.
The increase in the breadth and depth of sequence sampling provided by partial genomes comes with the caveat that they do not represent the entire proteome and are often biased toward highly expressed proteins. Nonetheless, previous reports support the legitimacy of these sequences, noting that findings from partial genome data sets are consistent with those from completed eukaryotic genomes (Brocchieri and Karlin 2005; Peregrin-Alvarez and Parkinson 2007). In comparing gene models derived from the recently published genome of P. vivax (Carlton et al. 2008), 69% were represented by an EST-contig (TBLASTN; E < 10−8) used in this study, further supporting the relevance of the findings.
The abundance of families located toward the terminal branches of the apicomplexan phylogeny reveals the relatively recent timings of large numbers of genetic adaptations acquired to mediate their diverse parasitic lifestyles. Interestingly, moving deeper within the apicomplexan phylogeny increased the number of protein families with meaningful functional annotations. This reflects either a greater emphasis placed by researchers on sequences found in more than one species, or a genuine difference in protein functions associated with different evolutionary origins. Among the families derived from the deeper nodes of the phylogeny were many examples of proteins generally perceived as being involved in conserved housekeeping functions, for example, ribosomal subunits (Supplemental Tables S1–S4). This is in contrast to previous studies of conserved nematode-specific families (Wasmuth et al. 2008). Whether this is truly reflective of apicomplexan-wide innovations or merely a consequence of Apicomplexa's ancient origins remains to be elucidated (Baldauf 2003). On the other hand, the large numbers of hypothetical proteins associated with the terminal branches suggest novel species- and/or genera-specific innovations.
This study has revealed remarkable sequence diversity associated with the Apicomplexa. It is striking that the major lineages (e.g., Cryptosporidium, Coccidia, Piroplasmida, and Haemosporidia) appear to share few parasite-associated proteins. Despite morphological and cytostructural similarities, these findings suggest that the emergence of the ancestral apicomplexan was followed by a potentially rapid evolution of parasitic strategies, each requiring their own battery of lineage-specific proteins. Changes in life cycle, in particular the use of vectors, has allowed the parasite to expand its host range (Jakes et al. 2003). This leads to strong selection pressures, resulting in a rapid and extensive modification of the proteome and frequent speciation. Consistent with the data presented here, it is likely that many lineage-specific protein families are associated with specialized parasite-related functions, including recognition of definitive host (for sexual recombination), survival in an insect vector, avoidance of immune responses, and transformation of the invaded cell to aid in the clonal replication of the parasite.
As noted earlier, the use of a less-stringent BLAST cut-off score can result in large-scale reclassification of taxon-restricted sequences (Table 2; Supplemental Fig. S4). Lower scoring matches are often indicative of local regions of sequence similarity typically representing protein domains. We therefore investigated the apicomplexan sequences from the perspective of protein domains. Considering the PfamA domain library, 52% of P. falciparum proteins can be annotated, which is less than many other complete eukaryote genomes, such as human (67%), Caenorhabditis elegans (64%), and Saccharomyces cerevisae (65%). PfamB coverage is higher for P. falciparum (68%) compared with model organisms (Hs 27%, Ce 42%, and Sc 53%), a consequence of P. falciparum sequence data not annotated by current PfamA models. Given the sequence divergence shown in this work, it seems likely that some known domains are present in the Apicomplexa, but have diverged from their Pfam domain profiles. Consequently, we suggest that many apicomplexan-specific PfamB-annotated domains may represent diverged members of existing PfamA domains.
The depth of protein space in the Apicomplexa is further expanded with the identification of novel domain combinations. As for the protein families, the incidence of these pairs occurred mainly at the level of species and genera (particularly when only PfamA domains are considered). We expect that further investigations of the novel domain pairings at other nodes within this phylogeny will yield additional insights underlying apicomplexan biology.
Conclusion
Here, we present an overview of the extent of sequence innovation within the Apicomplexa. Given the scale of this study, it has been possible to describe only a small selection of the more noteworthy examples of protein family and domain specializations. Within these data sets, however, we expect there to be many more important discoveries. Consequently, the data sets generated and discussed in this study are freely available on our project website at http://www.compsysbio.org/projects/apicomparison.
Methods
Sequences
Complete genomes were obtained from ApiDB (Aurrecoechea et al. 2007) (Plasmodium falciparum [v5.2], P. yoelii [v5.2], Cryptosporidium parvum [v3.4], and C. hominis [v3.4]) and GenBank, NCBI (Benson et al. 2005) (Theileria parva [April 2007] and T. annulata [April 2007]). The EST-contigs for the partial genomes were downloaded from dbEST (Boguski et al. 1993) on January 2007, processed, and assembled with the PartiGene suite (Parkinson et al. 2004) and are available from PartiGeneDB (Peregrin-Alvarez et al. 2005). EST-contigs were translated into peptide sequences with prot4EST (Wasmuth and Blaxter 2004).
Taxonomic profiles
BLAST searches (Altschul et al. 1997) were used to compare the apicomplexan sequences with UniProt (version 9.0—3,554,507 sequences) (Apweiler et al. 2004), COGENT (Feb 2007—915,554 sequences) (Janssen et al. 2003), and PartiGeneDB (Feb 2007—2,174,649 sequences) (Peregrin-Alvarez et al. 2005). For the BLAST programs (v2.2.13) used, see Supplemental Table S6.
Functional annotation
Sequences were annotated through sequence similarity comparisons and available high-throughput proteomic data. Proteomic data was available for P. falciparum (Florens et al. 2002), P. berghei (Hall et al. 2005), P. yoelii (Tarun et al. 2008), T. gondii (Bradley et al. 2005; Xia et al. 2008), and C. parvum (Sanderson et al. 2008). The data was downloaded from ApiDB (Aurrecoechea et al. 2007). We accepted proteins that were represented with a minimum of three spectra.
Annotation through sequence similarity involved three approaches: BLAST, COG/KOG, and Gene Ontology (see Supplemental File S1 for more details). These complemented information available in the description line for proteins from the published genomes. For all protein families that are described in the text, figures, and Supplemental Tables, the annotations were manually checked for inconsistencies.
Protein family generation
The two most commonly used tools for clustering protein sequences are TribeMCL (Enright et al. 2002) and OrthoMCL (Li et al. 2003b). OrthoMCL has been shown to be the best-performing method currently available when considering complete protein sets (Chen et al. 2007). As we are analyzing both complete and partial genomes, we chose to use TribeMCL, which does not explicitly model orthologous and paralogous relationships. An inflation parameter (I) of 3.0 was used. The protein family nomenclature is our own and composed of three elements: the “api” prefix shows that these families are from apicomplexans, the first number is the cluster designation, and the remaining number provides the Inflation parameter used for TribeMCL.
Apicomplexan phylogeny
Previous phylum-wide phylogenies had divided the Apicomplexa into three main taxa: the Coccidia, Haemosporidia, and Piroplasmida. The classification of Cryptosporidium as a coccidian has been revised and is now placed with the gregarines at the root of the Apicomplexa (Zhu et al. 2000a; Leander et al. 2003). In considering the phylogenetic relationships within the Apicomplexa and in the wider context of the Eukarya, studies from Zhu et al. (2000a) and Simpson and Roger (2004), respectively, were used.
Protein family alignments and phylogeny
Protein alignments were generated using the MUSCLE program (Edgar 2004) (default parameters). All alignments were inspected using Jalview (Clamp et al. 2004) and manually improved as necessary. Phylogenetic reconstructions were achieved using the PHYLIP suite (Felsenstein 2005). In each instance, 10,000 bootstrap replicates were used (seqboot; parameters—default). Maximum likelihood phylogenetic trees were built (proml; parameters—“S” set to no, all others default) and a consensus tree generation (consensus; parameters—majority rule extended).
Protein domain annotation
Both PfamA and PfamB domains (Finn et al. 2006) were assigned to the apicomplexan sequences. For PfamA sequences, the hidden Markov models (HMM) library was searched using HMMER v2.3.2 (http://hmmer.janelia.org/). For the complete genomes, the global domain model library (“ls”) was used and the curated gathering score (GA) cutoff was applied to each model. For the EST-derived partial genomes, we use a hybrid approach to maximize coverage (Wasmuth et al. 2008).
PfamB domains were identified using RPS-BLAST, as only the sequence alignments are available. Our criteria were that the alignment needed to cover at least 75% of the domain length. Overlapping annotations were sorted according to alignment score and the best scoring alignment was accepted.
Gene Ontology overrepresentation
Gene Ontology (GO) terms were assigned as described in the above section on functional annotation. The overrepresentation analysis was performed using Ontologizer (version 2.0) (Robinson et al. 2004). The “population” was the GO slim annotation of all apicomplexan protein domains. The “study” group was the protein domains that form apicomplexan-specific combinations. Term-for-Term (TFT) analysis was performed, with the Bonferroni multiple test correction.
Sushi domain analysis
Metazoan and viral Sushi domains were obtained from Pfam (Finn et al. 2006) (PF00084). All-against-all global pairwise alignments were performed using the EMBOSS implementation of the Needleman–Wunsch algorithm, needle (Needleman and Wunsch 1970; Rice et al. 2000). Cluster3.0 was used to hierarchically cluster the score matrix (Eisen et al. 1998). The scores were normalized around the mean and clustered using Euclidean distance and complete linkage. The clusters were viewed with TreeView (Saldanha 2004).
Acknowledgments
We thank our colleague Michael Grigg (National Institutes of Health, USA) for his help in improving the quality of the study and manuscript. Further gratitude goes to the three reviewers, whose comments and suggestions went a long way to strengthening this study. This work was funded by a grant from the Canadian Institutes of Health Research (CIHR #MOP84556) to J.P. and in part by a Research Training award from the Hospital for Sick Children to J.W. J.D. was supported by a MRes studentship from the Medical Research Council, UK. J.P. is supported by a CIHR New Investigators award and an Early Researcher award from the Ontario Ministry of Research and Innovation.
Footnotes
[Supplemental material is available online at www.genome.org and at www.compsysbio.org/projects/apicomparison.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083386.108.
References
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori Z, et al. The complete genome sequence of the apicomplexan. Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Ajioka JW, Boothroyd JC, Brunk BP, Heh A, Hillier L, Manger ID, Marra M, Overton GC, Roos DS, Wan KL, et al. Gene discovery by EST sequencing in Toxoplasma gondii reveals sequences restricted to the Apicomplexa. Genome Res. 1998;8:18–28. doi: 10.1101/gr.8.1.18. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinall TV, Joynson DH, Guy E, Hyde JE, Sims PF. The molecular basis of sulfonamide resistance in Toxoplasma gondii and implications for the clinical management of toxoplasmosis. J Infect Dis. 2002;185:1637–1643. doi: 10.1086/340577. [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C, Heiges M, Wang H, Wang Z, Fischer S, Rhodes P, Miller J, Kraemer E, Stoeckert CJ, Jr, Roos DS, et al. ApiDB: Integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 2007;35:D427–D430. doi: 10.1093/nar/gkl880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Bauer B, Engelbrecht S, Bakker-Grunwald T, Scholze H. Functional identification of alpha 1-giardin as an annexin of Giardia lamblia. FEMS Microbiol Lett. 1999;173:147–153. doi: 10.1111/j.1574-6968.1999.tb13496.x. [DOI] [PubMed] [Google Scholar]
- Baum J, Papenfuss AT, Baum B, Speed TP, Cowman AF. Regulation of apicomplexan actin-based motility. Nat Rev Microbiol. 2006;4:621–628. doi: 10.1038/nrmicro1465. [DOI] [PubMed] [Google Scholar]
- Belanger F, Derouin F, Grangeot-Keros L, Meyer L. Incidence and risk factors of toxoplasmosis in a cohort of human immunodeficiency virus-infected patients: 1988–1995. HEMOCO and SEROCO study groups. Clin Infect Dis. 1999;28:575–581. doi: 10.1086/515147. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST–database for “expressed sequence tags.”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, Dunn JD, Ferguson DJ, Sanderson SJ, Wastling JM, et al. Proteomic analysis of rhoptry organelles reveals many novel constituents for host–parasite interactions in Toxoplasma gondii. J Biol Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- Brayton KA, Lau AO, Herndon DR, Hannick L, Kappmeyer LS, Berens SJ, Bidwell SL, Brown WC, Crabtree J, Fadrosh D, et al. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 2007;3:1401–1413. doi: 10.1371/journal.ppat.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briquet S, Boschet C, Gissot M, Tissandie E, Sevilla E, Franetich JF, Thiery I, Hamid Z, Bourgouin C, Vaquero C. High-mobility-group box nuclear factors of Plasmodium falciparum. Eukaryot Cell. 2006;5:672–682. doi: 10.1128/EC.5.4.672-682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocchieri L, Karlin S. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 2005;33:3390–3400. doi: 10.1093/nar/gki615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Fuller AL, McDougald LR, Zhu G. Apicoplast genome of the coccidian Eimeria tenella. Gene. 2003;321:39–46. doi: 10.1016/j.gene.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiol Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciulla E, Emery A, Konz D, Krushkal J. Evolutionary history of orthopoxvirus proteins similar to human complement regulators. Gene. 2005;355:40–47. doi: 10.1016/j.gene.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Claudianos C, Dessens JT, Trueman HE, Arai M, Mendoza J, Butcher GA, Crompton T, Sinden RE. A malaria scavenger receptor-like protein essential for parasite development. Mol Microbiol. 2002;45:1473–1484. doi: 10.1046/j.1365-2958.2002.03118.x. [DOI] [PubMed] [Google Scholar]
- Cui L, Fan Q, Hu Y, Karamycheva SA, Quackenbush J, Khuntirat B, Sattabongkot J, Carlton JM. Gene discovery in Plasmodium vivax through sequencing of ESTs from mixed blood stages. Mol Biochem Parasitol. 2005;144:1–9. doi: 10.1016/j.molbiopara.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Daszak P, Ball SJ, Pittilo RM, Norton CC. Ultrastructural evidence for dense granule exocytosis by first-generation merozoites of Eimeria tenella in vivo. Parasitol Res. 1993;79:256–258. doi: 10.1007/BF00931902. [DOI] [PubMed] [Google Scholar]
- Dowse T, Soldati D. Host cell invasion by the apicomplexans: The significance of microneme protein proteolysis. Curr Opin Microbiol. 2004;7:388–396. doi: 10.1016/j.mib.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Recent advances in Neospora and neosporosis. Vet Parasitol. 1999;84:349–367. doi: 10.1016/s0304-4017(99)00044-8. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2705-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hajj H, Demey E, Poncet J, Lebrun M, Wu B, Galeotti N, Fourmaux MN, Mercereau-Puijalon O, Vial H, Labesse G, et al. The ROP2 family of Toxoplasma gondii rhoptry proteins: Proteomic and genomic characterization and molecular modeling. Proteomics. 2006;6:5773–5784. doi: 10.1002/pmic.200600187. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast NM, Kissinger JC, Roos DS, Keeling PJ. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol Biol Evol. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. University of Washington; Seattle, WA: 2005. PHYLIP (Phylogenetic Inference Package) version 3.6. Department of Genome Sciences. [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite. Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Bishop R, Shah T, de Villiers EP, Carlton JM, Hall N, Ren Q, Paulsen IT, Pain A, Berriman M, et al. Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science. 2005;309:134–137. doi: 10.1126/science.1110439. [DOI] [PubMed] [Google Scholar]
- Graat EA, Ploeger HW, Henken AM, De Vries Reilingh G, Noordhuizen JP, Van Beek PN. Effects of initial litter contamination level with Eimeria acervulina on population dynamics and production characteristics in broilers. Vet Parasitol. 1996;65:223–232. doi: 10.1016/s0304-4017(96)00952-1. [DOI] [PubMed] [Google Scholar]
- Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- Heintzelman MB, Mateer MJ. GpMyoF, a WD40 repeat-containing myosin associated with the myonemes of Gregarina polymorpha. J Parasitol. 2008;94:158–168. doi: 10.1645/GE-1339.1. [DOI] [PubMed] [Google Scholar]
- Howe DK. Initiation of a Sarcocystis neurona expressed sequence tag (EST) sequencing project: A preliminary report. Vet Parasitol. 2001;95:233–239. doi: 10.1016/s0304-4017(00)00418-0. [DOI] [PubMed] [Google Scholar]
- Huang J, Mullapudi N, Lancto CA, Scott M, Abrahamsen MS, Kissinger JC. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 2004;5:R88. doi: 10.1186/gb-2004-5-11-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Nichols G. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin Microbiol Rev. 2002;15:145–154. doi: 10.1128/CMR.15.1.145-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakes K, O'Donoghue PJ, Cameron SL. Phylogenetic relationships of Hepatozoon (Haemogregarina) boigae, Hepatozoon sp., Haemogregarina clelandi, and Haemoproteus chelodina from Australian reptiles to other Apicomplexa based on cladistic analyses of ultrastructural and life-cycle characters. Parasitology. 2003;126:555–559. [PubMed] [Google Scholar]
- Janssen P, Enright AJ, Audit B, Cases I, Goldovsky L, Harte N, Kunin V, Ouzounis CA. COmplete GENome Tracking (COGENT): A flexible data environment for computational genomics. Bioinformatics. 2003;19:1451–1452. doi: 10.1093/bioinformatics/btg161. [DOI] [PubMed] [Google Scholar]
- Jean L, Grosclaude J, Labbe M, Tomley F, Pery P. Differential localisation of an Eimeria tenella aspartyl proteinase during the infection process. Int J Parasitol. 2000;30:1099–1107. doi: 10.1016/s0020-7519(00)00099-0. [DOI] [PubMed] [Google Scholar]
- Kappe S, Bruderer T, Gantt S, Fujioka H, Nussenzweig V, Menard R. Conservation of a gliding motility and cell invasion machinery in Apicomplexan parasites. J Cell Biol. 1999;147:937–944. doi: 10.1083/jcb.147.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Unnasch N, Aiello DP. Sequence evidence for an altered genetic code in the Neospora caninum plastid. Int J Parasitol. 1999;29:1557–1562. doi: 10.1016/s0020-7519(99)00119-8. [DOI] [PubMed] [Google Scholar]
- Leander BS, Clopton RE, Keeling PJ. Phylogeny of gregarines (Apicomplexa) as inferred from small-subunit rDNA and beta-tubulin. Int J Syst Evol Microbiol. 2003;53:345–354. doi: 10.1099/ijs.0.02284-0. [DOI] [PubMed] [Google Scholar]
- Li L, Brunk BP, Kissinger JC, Pape D, Tang K, Cole RH, Martin J, Wylie T, Dante M, Fogarty SJ, et al. Gene discovery in the apicomplexa as revealed by EST sequencing and assembly of a comparative gene database. Genome Res. 2003a;13:443–454. doi: 10.1101/gr.693203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003b;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C, Vandepoele K, Van de Peer Y. Whole-genome analysis reveals molecular innovations and evolutionary transitions in chromalveolate species. Proc Natl Acad Sci. 2008;105:3427–3432. doi: 10.1073/pnas.0712248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier C, Adjogble KD, Daubener W, Delauw MF. Dense granules: Are they key organelles to help understand the parasitophorous vacuole of all apicomplexa parasites? Int J Parasitol. 2005;35:829–849. doi: 10.1016/j.ijpara.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- O'Keeffe AH, Green JL, Grainger M, Holder AA. A novel Sushi domain-containing protein of Plasmodium falciparum. Mol Biochem Parasitol. 2005;140:61–68. doi: 10.1016/j.molbiopara.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Pain A, Renauld H, Berriman M, Murphy L, Yeats CA, Weir W, Kerhornou A, Aslett M, Bishop R, Bouchier C, et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science. 2005;309:131–133. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- Parkinson J, Anthony A, Wasmuth J, Schmid R, Hedley A, Blaxter M. PartiGene–constructing partial genomes. Bioinformatics. 2004;20:1398–1404. doi: 10.1093/bioinformatics/bth101. [DOI] [PubMed] [Google Scholar]
- Peregrin-Alvarez JM, Parkinson J. The global landscape of sequence diversity. Genome Biol. 2007;8:R238. doi: 10.1186/gb-2007-8-11-r238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peregrin-Alvarez JM, Yam A, Sivakumar G, Parkinson J. PartiGeneDB–collating partial genomes. Nucleic Acids Res. 2005;33:D303–D307. doi: 10.1093/nar/gki109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrullo LA, Gallagher PJ, Elseviers D. The role of 2-methylthio-N6-isopentenyladenosine in readthrough and suppression of nonsense codons in Escherichia coli. Mol Gen Genet. 1983;190:289–294. doi: 10.1007/BF00330653. [DOI] [PubMed] [Google Scholar]
- Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Tropical infectious diseases: Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Robinson PN, Wollstein A, Bohme U, Beattie B. Ontologizing gene-expression microarray data: Characterizing clusters with Gene Ontology. Bioinformatics. 2004;20:979–981. doi: 10.1093/bioinformatics/bth040. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sanderson SJ, Xia D, Prieto H, Yates J, Heiges M, Kissinger JC, Bromley E, La K, Sinden RE, Tomley F, et al. Determining the protein repertoire of Cryptosporidium parvum sporozoites. Proteomics. 2008;8:1398–1414. doi: 10.1002/pmic.200700804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley LD. Intracellular parasite invasion strategies. Science. 2004;304:248–253. doi: 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- Simpson AG, Roger AJ. The real “kingdoms” of eukaryotes. Curr Biol. 2004;14:R693–R696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Soares DC, Gerloff DL, Syme NR, Coulson AF, Parkinson J, Barlow PN. Large-scale modelling as a route to multiple surface comparisons of the CCP module family. Protein Eng Des Sel. 2005;18:379–388. doi: 10.1093/protein/gzi039. [DOI] [PubMed] [Google Scholar]
- Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, Daly TM, Bergman LW, Kappe SH. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci. 2008;105:305–310. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton TJ, Iyer LM, Anantharaman V, Enomoto S, Abrahante JE, Subramanian GM, Hoffman SL, Abrahamsen MS, Aravind L. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14:1686–1695. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso MA, Omoto CK. Gregarina niphandrodes may lack both a plastid genome and organelle. J Eukaryot Microbiol. 2007;54:66–72. doi: 10.1111/j.1550-7408.2006.00229.x. [DOI] [PubMed] [Google Scholar]
- Trouiller P, Olliaro P, Torreele E, Orbinski J, Laing R, Ford N. Drug development for neglected diseases: A deficient market and a public-health policy failure. Lancet. 2002;359:2188–2194. doi: 10.1016/S0140-6736(02)09096-7. [DOI] [PubMed] [Google Scholar]
- Waller RF, McFadden GI. The apicoplast: A review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol. 2005;7:57–79. [PubMed] [Google Scholar]
- Wasmuth JD, Blaxter ML. prot4EST: Translating expressed sequence tags from neglected genomes. BMC Bioinformatics. 2004;5:187. doi: 10.1186/1471-2105-5-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth J, Schmid R, Hedley A, Blaxter M. On the extent and origins of genic novelty in the phylum Nematoda. PLoS Negl Trop Dis. 2008;2:e258. doi: 10.1371/journal.pntd.0000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NJ. Antimalarial drug resistance. J Clin Invest. 2004;113:1084–1092. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RJ, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Africa Malaria Report 2003. World Health Organization; Geneva, Switzerland: 2003. [Google Scholar]
- Wu Y, Wang X, Liu X, Wang Y. Data-mining approaches reveal hidden families of proteases in the genome of malaria parasite. Genome Res. 2003;13:601–616. doi: 10.1101/gr.913403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Sanderson SJ, Jones AR, Prieto JH, Yates JR, Bromley E, Tomley FM, Lal K, Sinden RE, Brunk BP, et al. The proteome of Toxoplasma gondii: Integration with the genome provides novel insights into gene expression and annotation. Genome Biol. 2008;9:R116. doi: 10.1186/gb.-2008-9-7-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Widme G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, et al. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, Hayden MS, Hieny S, Sutterwala FS, Flavell RA, Ghosh S, et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308:1626–1629. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- Zhu G, Keithly JS, Philippe H. What is the phylogenetic position of Cryptosporidium? Int J Syst Evol Microbiol. 2000a;50:1673–1681. doi: 10.1099/00207713-50-4-1673. [DOI] [PubMed] [Google Scholar]
- Zhu G, Marchewka MJ, Keithly JS. Cryptosporidium parvum appears to lack a plastid genome. Microbiology. 2000b;146:315–321. doi: 10.1099/00221287-146-2-315. [DOI] [PubMed] [Google Scholar]