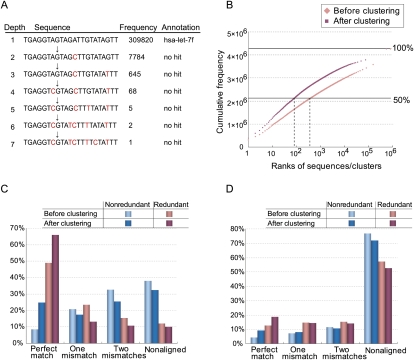
Figure 3.
Frequency-based de novo clustering improves short-read alignment to unique positions. Sequences are assigned to their parents if they fail in the nonsequencing error test (P < 0.01). (A) An example of the longest path in the cluster with the largest number of reads in the small RNAs sample. (B) Pink and red points indicate the cumulative frequencies of small RNAs ranked according to their frequencies before and after clustering, respectively. Their ranks are shown in the x-axis. (C) Percentages of redundant (or nonredundant) reads (or clusters) of the small RNAs sample that are aligned to the reference genome allowing for at most two mismatches. (D) Percentages of redundant (or nonredundant) 5′-end SAGE reads (or clusters) under the same conditions described in C.