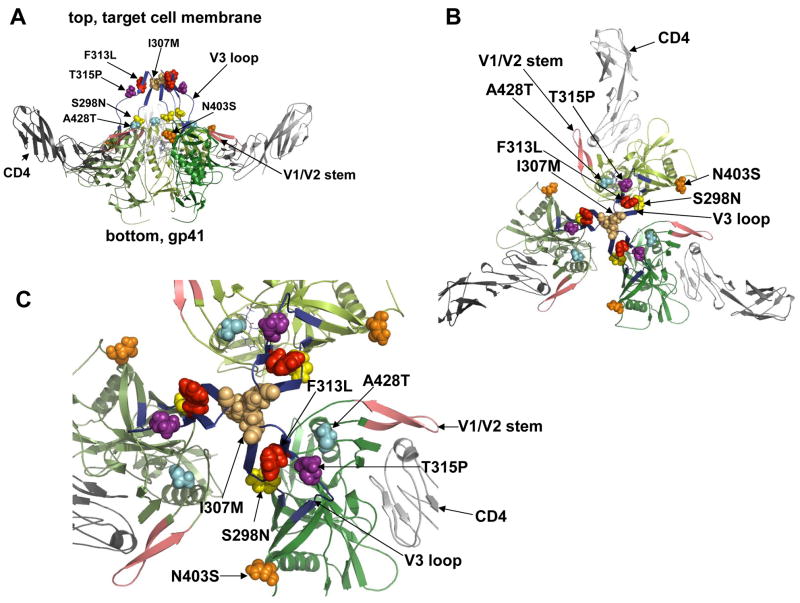
Fig 2.
Structural modeling of the trimeric gp120/CD4 complex showing the adaptive mutations as space-filling objects. The structure of HIV-1 JR-FL gp120 core protein containing the third variable region (V3) was used to generate the trimeric model in accordance with previous evidence. Frame A, side view of the gp120 trimer (green shaded) with associated CD4s (in grey) and indicating positions of the adaptive mutations near the top and gp41 at bottom. Frame B shows a top view in low magnification, with adaptive mutations clustered at the gp120 interfaces overlying the central channel above gp41. Frame C shows the top view at a higher magnification. This clustering of adaptive mutations supports the hypothesis that they collaboratively control allosteric changes in quaternary structure that prevent gp41 refolding.