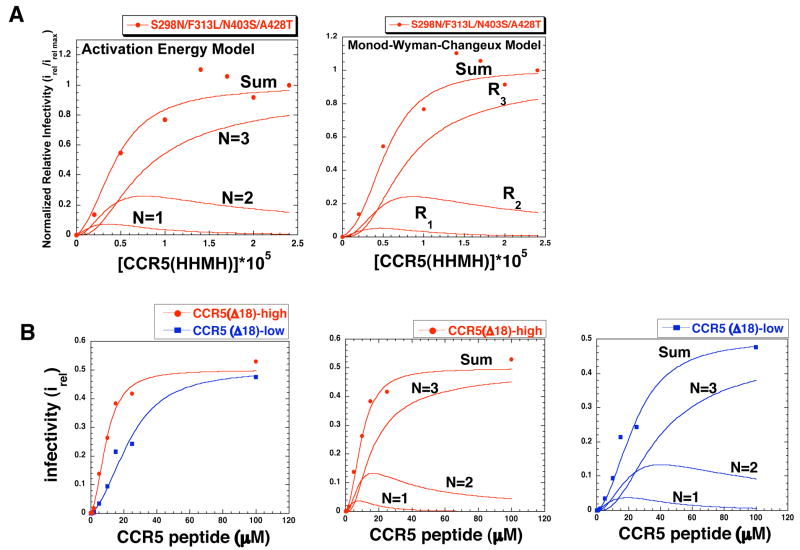
Fig 5.
Role of allostery in HIV-1 infections. (A, left panel) Activation Energy Model. The activation energy model (Eq 4) was fit to the infectivity data (Fig 4) for the virus with S298N/F313L/N403S/A428T that is highly adapted to CCR5(HHMH). (A, right panel) Monod-Wyman-Changeux Model. The latter model also fit to the same infectivity data. Both models closely fit the data and gave similar estimates for the contributions to infectivity of the viral complexes with different CCR5(HHMH) stoichiometries, of 1 (N=1, R1), 2 (N=2, R2), or 3 (N=3, R3). (B) Infection triggered by a tyrosine sulfated N-terminal CCR5 peptide. The N-terminal CCR5 peptide induced infections of HeLa-CD4 cells expressing low (2.7 × 104) or high (6.6 × 104) amounts of CCR5(Δ18) by the virus highly adapted to use CCR5(HHMH)-low. (B,left panel) Fit of infectivity data (irel values) using the activation energy allosteric model equation 4. The irel max values for both cell clones were 0.5. (B, middle and right panels) Deconvolution analysis of the peptide triggering data for cells with high (middle panel) and low (right panel) CCR5(Δ18), showing how complexes with 1, 2, or 3 peptides contributed to infection. Parameters obtained by fitting the data using equation 4 are given in Table 1.