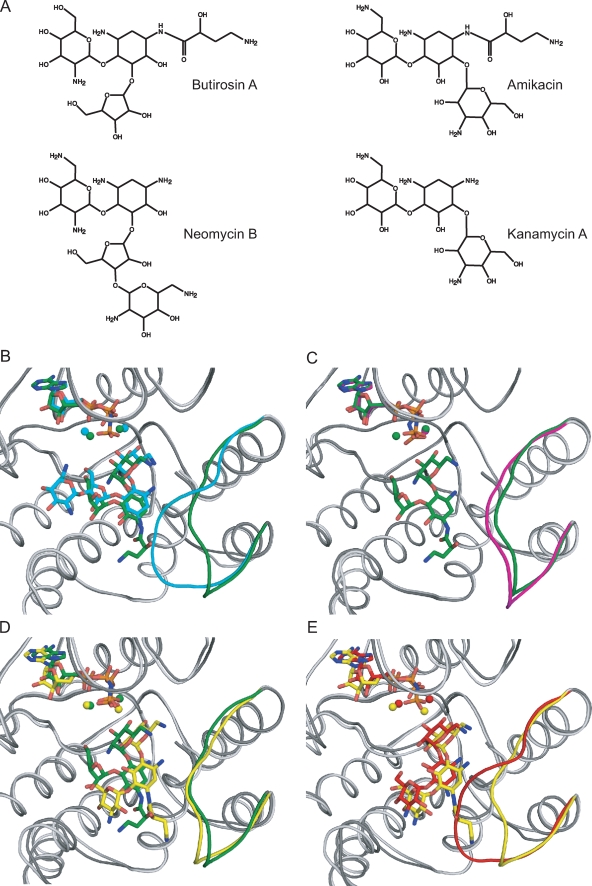
FIG. 3.
(A) Chemical structure of several aminoglycoside antibiotics. (B to E) Superpositions of APH(3′)-IIIa, shown in ribbon representation, bound with various ligands, drawn as sticks. The nucleotide, magnesium ions, aminoglycoside, and antibiotic-binding loops are highlighted in color. (B) APH(3′)-IIIa bound with butirosin A shown in dark gray/green is superposed on the neomycin B complex colored in light gray/cyan. (C) The butirosin A ternary complex is shown in dark gray/green, and the AMPPNP binary structure is shown in light gray/magenta. (D) The butirosin A-bound structure shown in dark gray/green is overlaid with the amikacin-bound model shown in light gray/yellow. (E) The kanamycin A ternary complex is shown in dark gray/red, and the APH(3′)-IIIa model bound with amikacin is shown in light gray/yellow. The RMSD value of the α-carbon atoms for residues 150 to 165 of the butirosin A ternary complex and the binary structure is 1.63 Å, whereas this value is 4.23 Å between the APH(3′)-IIIa-AMPPNP-butirosin A and APH(3′)-IIIa-ADP-kanamycin A or -neomycin B structures.