Abstract
Here we describe three Escherichia coli clinical isolates with reduced susceptibility to cefepime. Sequencing of the blaCMY genes revealed two novel variants (CMY-33 and -44) with two- to four-amino-acid deletions in the H-10 helix. The deletions were responsible for 12- to 24-fold increases in the MICs of cefepime.
CMY-2 is the most commonly encountered plasmid-mediated class C β-lactamase, often found in Escherichia coli and Salmonella enterica serovars (10, 13). The spectrum of resistance conferred by CMY-2 is typical of class C enzymes: CMY-2 confers resistance to penicillins, cephalosporins (including cephamycins), and aztreonam. In contrast, gram-negative bacteria harboring CMY-2 are susceptible to cefepime. Cefepime, a cephalosporin with methoxyimino and aminothiazolyl moieties, is a substrate that is stable against hydrolysis by most class C β-lactamases, both chromosomal and plasmid mediated. Here, we report the identification of two variants of CMY-2 β-lactamases, designated CMY-33 and CMY-44, which confer reduced susceptibility to cefepime.
E. coli strains producing these variants were isolated from three unrelated patients who were admitted to the University of Pittsburgh Medical Center (UPMC) Presbyterian Campus between 2006 and 2008. The E. coli strains under study were bloodstream and urinary tract isolates. Two of the patients received cefepime for 6 to 10 days within the 2 months preceding the isolation of the organisms. The MICs of the representative β-lactams for these clinical isolates were measured by Etest (AB Biodisk, Solna, Sweden). According to the Etest results, the E. coli isolates were all highly resistant to oxyiminocephalosporins, including cefuroxime, ceftazidime, and cefotaxime, and also showed reduced susceptibility to cefepime (MIC range, 6 to 96 μg/ml) (Table 1). Analytical isoelectric focusing revealed β-lactamase activity with pIs of >9.0 for all isolates and 5.4 for isolates YD006 and 34943, suggesting the presence of class C β-lactamases and TEM-1, respectively (data not shown). To define the β-lactamases causing the extended-spectrum cephalosporin resistance, PCR and sequencing were performed to detect the blaTEM-, blaSHV-, and blaCTX-M-type extended-spectrum β-lactamase (ESBL) genes (4). For the amplification and sequencing of the blaCMY-2-type β-lactamase genes, the following primer set was used: CMY-flanking-F (5′-CCGGACACCTTTTTGCTTTT-3′) and CMY-flanking-R (5′-TATCCTGGGCCTCATCGTCAGTTA-3′). The PCR amplification identified blaTEM-1 in isolates YD006 and 34943 but no class A ESBL genes in any of the isolates. In contrast, the PCR amplified blaCMY in all three isolates. The conjugal transfer of the blaCMY gene using E. coli J53 Azir (azide resistant) as the recipient (kind gift from George A. Jacoby) was successful with E. coli 34943 but not with E. coli 1285 or E. coli YD006. However, a plasmid was successfully transformed into E. coli DH10B by electroporation for all three isolates, indicating the plasmidic location of these blaCMY-2 variants. These plasmids appeared unrelated to each other by restriction endonuclease digestion (Fig. 1). The plasmid from E. coli YD006 conferred resistance to tetracycline, chloramphenicol, and sulfisoxazole in addition to cephalosporins; resistance to aminoglycosides was not observed. The plasmids from E. coli 1285 and 34943 only conferred resistance to cephalosporins. Sequencing analysis revealed the presence of novel CMY-2 variants, blaCMY-33 in isolates 1285 and YD006 and blaCMY-44 in isolate 34943, respectively. A two-amino-acid deletion (Leu293 and Ala294) was detected in the translated sequence of blaCMY-33 compared with that of the other CMY-2 variant. The translated sequence of blaCMY-44 had an additional two-amino-acid deletion of Ala295 and Leu296 compared with that of blaCMY-33 (Fig. 2). These deletions were located within or in the proximity of the H-10 helix of the enzymes (alternatively, referred to as the R2 loop) (7).
TABLE 1.
Susceptibility of E. coli strains to representative β-lactams
| β-Lactam | MIC (μg/ml) for E. coli strain:
|
||||||
|---|---|---|---|---|---|---|---|
| 1285 (CMY-33) | YD006 (CMY-33) | 34943 (CMY-44) | DH10B (pCMY-33)a | DH10B (pCMY-44)a | DH10B (pCMY-2)a | DH10B (pBCSK−) | |
| Ampicillin | >256 | 128 | >256 | >256 | >256 | >256 | 3 |
| Piperacillin | >256 | 12 | >256 | 64 | 32 | 128 | 2 |
| Piperacillin-tazobactam | 128 | 2 | 96 | 6 | 16 | 4 | 1 |
| Cefuroxime | >256 | 96 | >256 | 128 | 64 | >256 | 6 |
| Ceftazidime | >256 | >256 | >256 | >256 | >256 | >256 | 0.5 |
| Cefotaxime | >256 | 8 | 32 | 12 | 8 | 16 | 0.064 |
| Cefoxitin | >256 | 24 | >256 | 24 | 32 | >256 | 6 |
| Cefepime | 96 | 6 | 32 | 6 | 3 | 0.25 | 0.032 |
| Aztreonam | 64 | 6 | 24 | 16 | 4 | 8 | 0.064 |
| Ertapenem | 1 | 0.064 | 4 | 0.032 | 0.064 | 0.094 | 0.012 |
Isogenic strains of E. coli DH10B with pBCSK− vector containing blaCMY-33, blaCMY-44, and blaCMY-2, respectively.
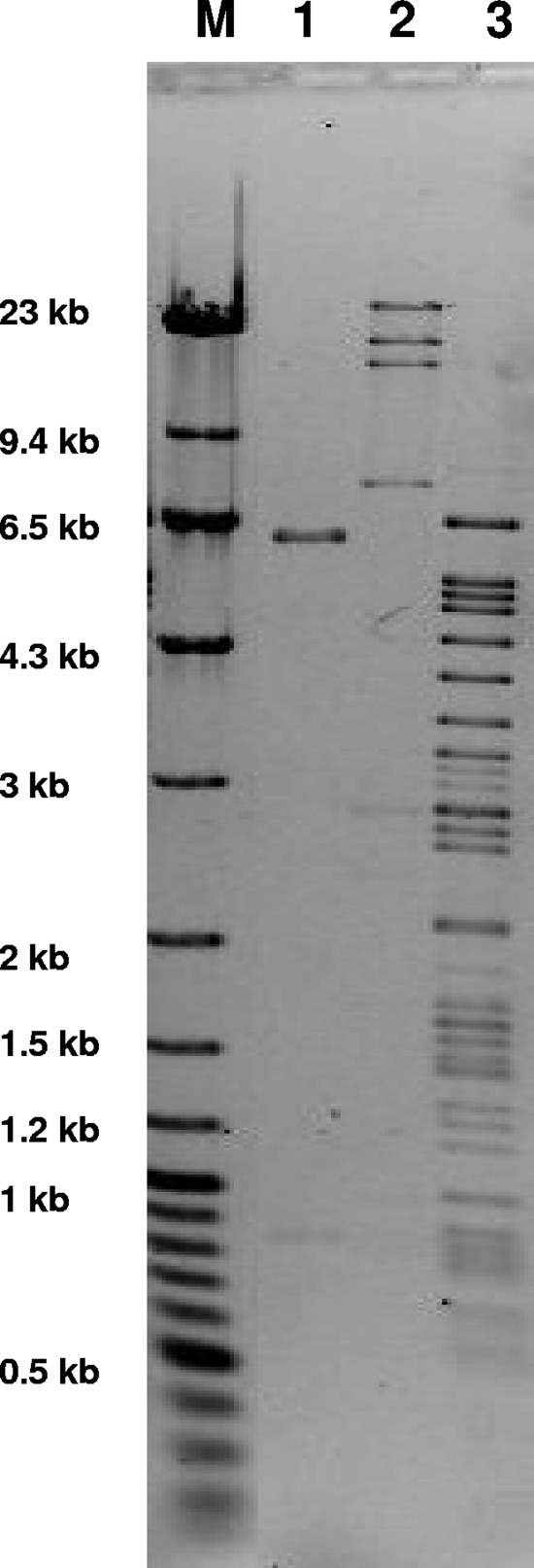
FIG. 1.
Plasmid restriction analysis of the blaCMY-bearing plasmids in E. coli DH10B transformants corresponding to the clinical isolates. Lane M, molecular weight markers (λ DNA-HindIII digest [New England Biolabs, Ipswich, MA] plus GeneRuler 100-bp Plus DNA ladder [Fermentas, Glen Burnie, MD]); lane 1, 1285; lane 2, YD006; lane 3, 34943. The plasmids were extracted by the standard alkaline lysis method, digested with PstI (New England Biolabs), and electrophoresed in a 0.7% agarose gel.
FIG. 2.
A sequence alignment of amino acid residues in the H-10 helix region of class C β-lactamases. Partial amino acid sequences of CMY-33 (GenBank accession no. EU496816), CMY-43 (GenBank accession no. FJ437066), and CMY-2 (GenBank accession no. X91840) are aligned along with AmpC β-lactamases of Escherichia coli HKY28 (AB108683), Enterobacter cloacae CHE (AJ278994), and Serratia marcescens HD (AY336102). Amino acids within the R2 loop are represented by a square box. The numbering scheme of the amino acid residues of P99 is adopted (6).
In class C β-lactamases, certain amino acid deletions in the H-10 helix are implicated in the broadening of the substrate spectrum to include cefepime (2, 5, 8). To establish the role of these deletions in the reduced susceptibility to cefepime, isogenic E. coli strains producing CMY-2, CMY-33, or CMY-44 were constructed. In brief, the three blaCMY genes were amplified by PCR and directionally cloned to vector pBCSK− as described previously (3). E. coli DH10B was then transformed with these recombinant plasmids. The MICs against representative β-lactams are presented in Table 1.
The E. coli DH10B strains producing CMY-33 and CMY-44 demonstrated reduced susceptibility to cefepime compared with that producing CMY-2 by 12- to 24-fold, whereas resistance to ceftazidime and cefotaxime was maintained (Table 1). On the other hand, the MICs of cefuroxime and cefoxitin, which are usually excellent substrates of class C β-lactamases including CMY-2, were lowered for isolates producing CMY-33 and -44. These results suggested that the amino acid deletions observed in CMY-33 and -44 may be responsible for the reduced cefepime susceptibility in these clinical isolates. Given the variable degree of resistance to cefepime among the clinical isolates, however, it is plausible that other mechanisms augmenting cefepime resistance are present, especially in isolate 1285.
Certain amino acid changes (insertions, deletions, or substitutions) in the H-10 helix are believed to result in structural alterations that enable better accommodation of the R2 side chains of cephalosporins, including cefepime, into the active site of class C β-lactamases and thus enhance the hydrolysis of these substrates (7). Examples of insertions and substitutions in class C enzymes that lead to a clinically significant reduction of cefepime susceptibility include chromosomal AmpC of E. coli BER (9) and Enterobacter aerogenes Ear2 (1) and plasmid-mediated CMY-19 in Klebsiella pneumoniae (12). Examples of deletions leading to this property include chromosomal AmpC of Enterobacter cloacae CHE (2), Serratia marcescens HD (8), and E. coli HKY28 (5). All of these variant enzymes were reported as sporadic events, and the understanding is that cefepime maintains excellent activity against organisms producing class C β-lactamases (11). However, our finding of three independent clinical cases producing two CMY-2 variants with this property during a relatively brief period of time suggests that blaCMY-2, the most common plasmid-mediated class C β-lactamase gene in E. coli, has the potential to develop variants with reduced susceptibility to cefepime, likely under the selective pressure from excessive use. Clinical microbiologists should be aware that reduced susceptibility to cefepime may occur in variants of CMY-2, which are not easily identified in the clinical laboratory and thus may be overlooked. The continuing discovery of plasmid-mediated AmpCs has significant implications for infection control measures.
Nucleotide sequence accession numbers.
The sequences determined in this study appear in the GenBank/EMBL/DDBJ database under accession numbers EU496816 and FJ437066.
Acknowledgments
Part of this work was supported by a joint fellowship grant from the National Foundation of Infectious Diseases and the Infectious Diseases Society of America. Y.D. was supported by an NIH training grant (T32 AI007333). D.L.P. is supported in part by the NIH (R01 AI070896). R.A.B. is supported by the NIH (R01 AI063517), Merit Review Award, and the Geriatric Research Education and Clinical Center (GRECC), VISN 10. A.E. is supported by grants from AstraZeneca and the NIH (R01 AI063517).
Footnotes
Published ahead of print on 4 May 2009.
REFERENCES
- 1.Barnaud, G., Y. Benzerara, J. Gravisse, L. Raskine, M. J. Sanson-Le Pors, R. Labia, and G. Arlet. 2004. Selection during cefepime treatment of a new cephalosporinase variant with extended-spectrum resistance to cefepime in an Enterobacter aerogenes clinical isolate. Antimicrob. Agents Chemother. 48:1040-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnaud, G., R. Labia, L. Raskine, M. J. Sanson-Le Pors, A. Philippon, and G. Arlet. 2001. Extension of resistance to cefepime and cefpirome associated to a six-amino-acid deletion in the H-10 helix of the cephalosporinase of an Enterobacter cloacae clinical isolate. FEMS Microbiol. Lett. 195:185-190. [DOI] [PubMed] [Google Scholar]
- 3.D'Andrea, M. M., E. Nucleo, F. Luzzaro, T. Giani, R. Migliavacca, F. Vailati, V. Kroumova, L. Pagani, and G. M. Rossolini. 2006. CMY-16, a novel acquired AmpC-type β-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from northern Italy. Antimicrob. Agents Chemother. 50:618-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Oliveira Garcia, D., Y. Doi, D. Szabo, J. M. Adams-Haduch, T. M. I. Vaz, D. Leite, M. C. Padoveze, M. P. Freire, F. P. Silveira, and D. L. Paterson. 2008. Multiclonal outbreak of Klebsiella pneumoniae producing extended-spectrum β-lactamase CTX-M-2 and novel variant CTX-M-59 in a neonatal intensive care unit in Brazil. Antimicrob. Agents Chemother. 52:1790-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi, Y., J. Wachino, M. Ishiguro, H. Kurokawa, K. Yamane, N. Shibata, K. Shibayama, K. Yokoyama, H. Kato, T. Yagi, and Y. Arakawa. 2004. Inhibitor-sensitive AmpC β-lactamase variant produced by an Escherichia coli clinical isolate resistant to oxyiminocephalosporins and cephamycins. Antimicrob. Agents Chemother. 48:2652-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galleni, M., F. Lindberg, S. Normark, S. Cole, N. Honore, B. Joris, and J. M. Frere. 1988. Sequence and comparative analysis of three Enterobacter cloacae ampC beta-lactamase genes and their products. Biochem. J. 250:753-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, J. Y., H. I. Jung, Y. J. An, J. H. Lee, S. J. Kim, S. H. Jeong, K. J. Lee, P. G. Suh, H. S. Lee, S. H. Lee, and S. S. Cha. 2006. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C β-lactamase. Mol. Microbiol. 60:907-916. [DOI] [PubMed] [Google Scholar]
- 8.Mammeri, H., L. Poirel, P. Bemer, H. Drugeon, and P. Nordmann. 2004. Resistance to cefepime and cefpirome due to a 4-amino-acid deletion in the chromosome-encoded AmpC β-lactamase of a Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 48:716-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mammeri, H., L. Poirel, and P. Nordmann. 2007. Extension of the hydrolysis spectrum of AmpC β-lactamase of Escherichia coli due to amino acid insertion in the H-10 helix. J. Antimicrob. Chemother. 60:490-494. [DOI] [PubMed] [Google Scholar]
- 10.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenover, F. C., S. L. Emery, C. A. Spiegel, P. A. Bradford, S. Eells, A. Endimiani, R. A. Bonomo, and J. E. McGowan, Jr. 2009. Identification of plasmid-mediated AmpC β-lactamases in Escherichia coli, Klebsiella spp., and Proteus species can potentially improve reporting of cephalosporin susceptibility testing results. J. Clin. Microbiol. 47:294-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wachino, J., H. Kurokawa, S. Suzuki, K. Yamane, N. Shibata, K. Kimura, Y. Ike, and Y. Arakawa. 2006. Horizontal transfer of blaCMY-bearing plasmids among clinical Escherichia coli and Klebsiella pneumoniae isolates and emergence of cefepime-hydrolyzing CMY-19. Antimicrob. Agents Chemother. 50:534-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walther-Rasmussen, J., and N. Hoiby. 2002. Plasmid-borne AmpC β-lactamases. Can. J. Microbiol. 48:479-493. [DOI] [PubMed] [Google Scholar]