Abstract
During phenotypic characterization of various Escherichia coli mutants, we observed that ΔphoA strains are capable of using glycerol-2-phosphate (G2P) as a sole source of phosphorus. Mutations in the ugpBAECQ operon eliminated this phenotype, suggesting that G2P is a previously unrecognized substrate for the binding protein-dependent Ugp transporter.
Most organisms preferentially use inorganic phosphate as a source of phosphorus. Nevertheless, ecosystems are commonly limited by this essential nutrient, and living organisms frequently encounter phosphorus starvation conditions. To overcome this problem, many microorganisms have evolved systems for the acquisition of phosphorus from alternative substrates, which include a wide variety of phosphate esters, phosphonic acids and other reduced phosphorus compounds (17, 19).
There are two generic mechanisms for utilization of phosphate esters as a source of phosphorus. Many phosphate esters are hydrolyzed by extracellular phosphatases that release free phosphate, which is then transported into the cell. Other phosphate esters are transported intact and then either hydrolyzed by a cytoplasmic phosphatase or, in cases where the compound is a normal cellular metabolite, channeled directly into metabolism. The use of these alternate phosphorus sources in the enteric bacterium Escherichia coli has been intensively studied (for a review, see reference 17). In this organism the principal extracellular phosphatase is a nonspecific, alkaline phosphomonoesterase encoded by the phoA gene (4). PhoA is required for the use of a variety of phosphate esters as sole phosphorus sources; however, glycerol-3-phosphate (G3P) and G3P diesters can serve as sole phosphorus sources in the absence of PhoA after uptake by a binding protein-dependent transporter encoded by the ugpBAECQ operon (2, 12). The last gene in the operon, ugpQ, encodes a glycerol-3-phosphodiesterase that releases free G3P after transport (2). Consistent with their proposed biological roles, both ugp and phoA are tightly regulated by the availability of extracellular phosphate, along with a number of other genes involved in the use of alternative phosphorus sources. These genes, collectively known as the PHO regulon, are transcriptionally controlled by the products of the phoBR operons and are expressed only during phosphate starvation (15).
Gylcerol-2-phosphate (G2P), on the other hand, has been presumed to be a nontransportable substrate and, thus, dependent on PhoA for its use as a source of phosphorus. In fact, several publications have reported the use of G2P for selection for alkaline phosphatase mutants (9, 11, 14). Therefore, we were surprised to find that a ΔphoA strain (BW14894 [Table 1]) was able to utilize G2P (Sigma, St. Louis, MO) as a sole phosphorus source in 0.4% glucose-morpholinepropanesulfonic acid (MOPS) medium (18). We considered two possible explanations for this result.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BW14893 | Δlac(X74) ΔphoA532 Δphn33-30 | 8 |
| BW14894 | Δlac(X74) Δphn33-30 | 20 |
| WM5085 | Δlac(X74) ΔphoA532 Δphn33-30 ΔugpBAECQ | This study |
| WM5089 | Δlac(X74) ΔphoA532 Δphn33-30 ΔugpB | This study |
| WM5063 | Δlac(X74) ΔphoA532 Δphn33-30 ΔugpC | This study |
| WM5086 | Δlac(X74) ΔphoA532 Δphn33-30 ΔugpQ | This study |
| WM5087 | Δlac(X74) ΔphoA532 Δphn33-30 ΔugpE | This study |
| WM5088 | Δlac(X74) ΔphoA532 Δphn33-30 ΔugpA | This study |
| WM7184 | Δlac(X74) ΔphoA532 Δphn33-30 ΔglpT::aph | This study |
| WM7186 | Δlac(X74) ΔphoA532 Δphn33-30 ΔglpD::aph | This study |
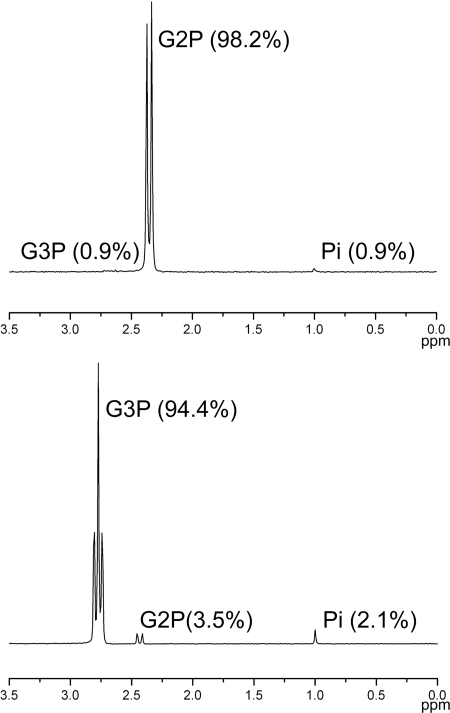
First, it seemed possible that our G2P stock solution was contaminated by another phosphorus compound, such as G3P, that could serve as a sole phosphorus source in the absence of PhoA. To test this hypothesis, we analyzed 500 mM stock solutions of G2P and G3P (Sigma, St. Loius, MO) using 31P nuclear magnetic resonance (NMR) as previously described (21). Proton-coupled 31P spectra of G2P showed the expected doublet with peaks at 2.38 and 2.32 ppm, consistent with previously reported chemical shifts (3) (Fig. 1). Very small additional peaks were observed at 2.7 and 1.0 ppm, which were consistent with minor amounts (less than 1%) of G3P and phosphate, respectively, in the sample. Spectra of G3P show the predicted triplet centered at 2.8 ppm (3), accounting for 98% of the phosphorus present with 3.5% G2P and 2.1% phosphate. Control experiments conducted by spiking samples with the corresponding source at a concentration of 5% showed that additional contaminating phosphorus compounds would have been easily detected at this level; therefore, no additional phosphorus compounds were present (data not shown).
FIG. 1.
Proton-coupled 31P NMR spectra for G2P and G3P. Solutions (500 mM) of the highest grades commercially available were analyzed by 31P NMR as described in the text. The spectrum of G2P has the predicted doublet at 2.38 and 2.32 ppm, with splitting caused by the single proton on the C-2 position. The spectrum of G3P has the predicted triplet centered at 2.80 ppm, with splitting caused by two protons at the C-3 position. Integration of the observed peaks shows that G2P is contaminated by minor amounts of phosphate (Pi) and G3P, whereas G3P has slightly higher concentrations of both phosphate and G2P. The percentage of the total phosphorus for each compound is indicated in parentheses. The spectra were aligned using the phosphate peak at 1 ppm.
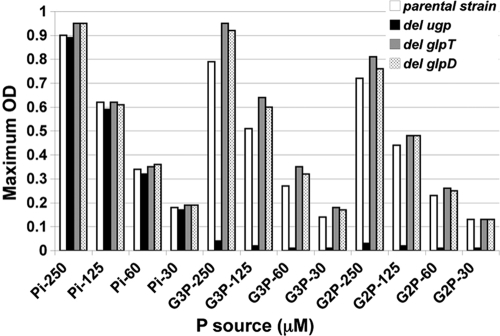
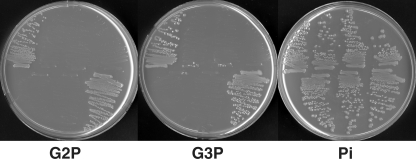
The second possibility that we considered was that our strain had a previously unrecognized phoA-independent pathway for assimilation of phosphorus from G2P. Because PhoA is the predominant extracellular phosphatase in E. coli, such a pathway would likely entail transport of G2P. Two binding protein-dependent transport systems have been shown to allow use of phosphate esters as a source of phosphorus in E. coli: the ugp system described above and the broad-specificity phnCDE system, which allows uptake of phosphonates, phosphate esters, and inorganic phosphate (10). E. coli also possesses a second G3P transporter, encoded by glpT, which can allow use of G3P as a phosphorus source under certain circumstances (6). The strains used in this study all have a deletion of the phn operon, ruling out involvement of this transport system. To test whether the Ugp transporter was involved, we constructed a series of deletion mutants lacking each of the ugp genes and lacking the entire ugp operon. The mutations were constructed using E. coli BW26678 by λ Red-mediated recombination as described previously (5), using the kanamycin resistance marker on pKD13 and the oligonucleotides shown in Table 2. Each mutation was then moved into BW14894 by P1kc-mediated phage transduction as described previously (16). Finally, the kanamycin resistance cassette was removed by flp-mediated recombination to generate nonpolar deletion mutations (5). A glpT mutant was constructed in a similar fashion, but the kanamycin resistance marker, which was derived from pKD4, was not removed. The final strains are shown in Table 1. The abilities of the mutants to utilize G2P, G3P, and phosphate as sole phosphorus sources were examined by scoring growth in 0.4% glucose-MOPS media with various growth-limiting concentrations of the phosphorus sources (Fig. 2). Mutants with glpT deletions were able to utilize both G2P and G3P as phosphorus sources, showing that glpT is not required for this process. In contrast, mutants lacking ugpBEACQ failed to grow on both G2P and G3P. Similar results were obtained with ugpB, ugpE, ugpA, and ugpC mutants (Fig. 3). Thus, in this ΔphoA strain background, growth on G2P and G3P requires the Ugp transporter. The phosphodiesterase-encoding gene ugpQ is not required for use of either compound. The observation that the growth on each substrate (in cases where growth occurred) was proportional to the level of phosphorus added and equivalent to the growth obtained with the same concentration of inorganic phosphate further corroborates the conclusion that growth was dependent on the specific phosphorus compound used and not due to contaminating compounds present in our source chemicals.
TABLE 2.
Oligonucleotide primers used for deletion constructiona
| Deletion | Direction | Primer |
|---|---|---|
| ugpBAECQ | Forward | ATGAAACCGTTACATTATACAGCTTCAGCACTGGCGGTGTAGGCTGGAGCTGCTTCG |
| Reverse | CTATTGGGCCGTAAAGTTCGGACCAATCACGTCAATTTCCGGGGATCCGTCGACCTG | |
| ugpB | Forward | ATGAAACCGTTACATTATACAGCTTCAGCACTGGCGGTGTAGGCTGCAGCTGCTTCG |
| Reverse | TTAAGACTTCGTCGATTTCTCAAAGCGGCGCAGCAATTCCGGGGATCCGTCGACCTG | |
| ugpE | Forward | ATGATTGAGAACCGTCCGTGGCTGACGATATTCAGCGTGTAGGCTGGAGCTGCTTCG |
| Reverse | TTATTTCTCACTATCGACCAGGCCGCGCGCGAAGGCTTCCGGGGATCCGTCGACCTG | |
| ugpA | Forward | ATGTCATCATCCCGTCCGGTGTTCCGCTCGCGCTGGGTGTAGGCTGGAGCTGCTTCG |
| Reverse | TCATTGGTAACGCACCTTGCTTTCAACATAGCGGAATTCCGGGGATCCGTCGACCTG | |
| ugpC | Forward | ATGGCAGGACTGAAATTACAGGCAGTAACCAAAAGCGTGTAGGCTGGAGCTGCTTCG |
| Reverse | TCATACTCGTTGTCCTGTTTCACCATCAAAAAGATGTTCCGGGGATCCGTCGACCTG | |
| ugpQ | Forward | ATGAGTAACTGGCCTTATCCCCGCATCGTCGCTCATGTGTAGGCTGGAGCTGCTTCG |
| Reverse | CTATTGGGCCGTAAAGTTCGGACCAATCACGTCAATTCCGGGGATCCGTCGACCTG | |
| glpT | Forward | CATGAACAATTACTGCAAGAACGCAACGGAGGCTAATGGCGTGTAGGCTGGAGCTGCTTC |
| Reverse | TCATCATGATCGCCATGCTAAGGTTTTTCAGCGTCAATTTCATATGAATATCCTCCTTAG | |
| glpD | Forward | CGAACATTTATGAGCTTTAACGAAAGTGAATGAGGGCAGCGTGTAGGCTGGAGCTGCTTC |
| Reverse | CAGGCCAGATTGAAATCTGACCTGATCACCTTACGTTAATCATATGAATATCCTCCTTAG |
Bold type indicates sequences that are homologous to regions adjacent to the genes indicated; the remainder of each primer is homologous to pKD13 or pKD4 and primes amplification of the kanamycin resistance cassette (5).
FIG. 2.
Growth yields for selected E. coli mutants in media with G2P, G3P, and phosphate (Pi) as sole phosphorus sources. The growth yield was determined by measuring the final optical density at 600 nm (OD) after growth in 0.4% glucose-MOPS liquid media with various concentrations of the phosphorus sources. All strains are derivatives of BW14893 [Δlac(X74) ΔphoA532 Δphn33-30]. The strains tested were BW14893 (parental strain), WM5085 (del ugp), WM7184 (del glpT), and WM7186 (del glpD). The concentrations used were 30, 60, 125, and 250 μM.
FIG. 3.
Utilization of G2P, G3P, and phosphate (Pi) as sole phosphorus sources by individual ugp mutants. Strains were streaked on 0.4% glucose-MOPS media with the phosphorus sources indicated at a concentration of 0.5 mM. All strains are derivatives of BW14893 [Δlac(X74) ΔphoA532 Δphn33-30]. The following strains were tested (from left to right): top row, BW14893, WM5085 (ΔugpBAECQ), WM5089 (ΔugpB), and WM5087 (ΔugpE); bottom row, WM5088 (ΔugpA), WM5063 (ΔugpC), and WM5086 (ΔugpQ).
It is formally possible that G2P is enzymatically rearranged to form G3P prior to uptake by Ugp. We attempted to address this issue by introducing a mutation that would block posttransport processing of G3P. Unfortunately, the enzyme(s) that mediates assimilation of P from cytoplasmic G3P is unknown. It is known that the product of the glpD gene, glycerol-3-phosphate dehydrogenase, is required for use of G3P as a carbon source (13). Therefore, we constructed a glpD mutant (as described above for the glpT mutant) and tested its ability to utilize G2P and G3P as sole phosphorus sources. As shown in Fig. 2, the glpD mutation had no effect on the use of either phosphorus source. Thus, the possibility of conversion of G2P to G3P remains open, as does the downstream fate of G3P following transport by Ugp.
The requirement for an intact Ugp transporter for use of G2P strongly suggests that this compound is a previously unrecognized substrate for this binding protein-dependent uptake system. The Ugp system is already known to possess fairly broad specificity, since it is able to transport both G3P and a range G3P phosphodiesters (2). Further, in vitro studies with the single-subunit GlpT transporter have indicated that it can transport both G2P and G3P, providing a precedent for recognition of both molecules by a G3P uptake system (1). In contrast to G3P, which is a major component of phospholipids, G2P is not commonly thought of as a natural product. Nevertheless, recent studies have shown that G2P is produced by certain organisms (7) and hence should be considered an available phosphorus source in nature. Thus, a plausible selective pressure for evolution of G2P transport can also be envisioned. Further studies are required to determine the intracellular pathways for assimilation of phosphorus from both G2P and G3P.
Acknowledgments
This work was supported by grant GM059334B from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 8 May 2009.
REFERENCES
- 1.Auer, M., M. J. Kim, M. J. Lemieux, A. Villa, J. Song, X. D. Li, and D. N. Wang. 2001. High-yield expression and functional analysis of Escherichia coli glycerol-3-phosphate transporter. Biochemistry 406628-6635. [DOI] [PubMed] [Google Scholar]
- 2.Brzoska, P., and W. Boos. 1988. Characteristics of a ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the ugp transport system of Escherichia coli. J. Bacteriol. 1704125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buenemann, E. K., R. J. Smernik, A. L. Doolette, P. Marschner, R. Stonor, S. A. Wakelin, and A. M. McNeill. 2008. Forms of phosphorus in bacteria and fungi isolated from two Australian soils. Soil. Biol. Biochem. 401908-1915. [Google Scholar]
- 4.Coleman, J. E. 1992. Structure and mechanism of alkaline phosphatase. Annu. Rev. Biophys. Biomol. Struct. 21441-483. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi, S., J. P. Koch, and E. C. Lin. 1964. Active transport of l-α-glycerophosphate in Escherichia coli. J. Biol. Chem. 2393098-3105. [PubMed] [Google Scholar]
- 7.Jones, C., and X. Lemercinier. 2005. Full NMR assignment and revised structure for the capsular polysaccharide from Streptococcus pneumoniae type 15B. Carbohydr. Res. 340403-409. [DOI] [PubMed] [Google Scholar]
- 8.Lee, K. S., W. W. Metcalf, and B. L. Wanner. 1992. Evidence for two phosphonate degradative pathways in Enterobacter aerogenes. J. Bacteriol. 1742501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levinthal, C. 1959. Genetic and chemical studies with alkaline phosphatase of E. coli. Brookhaven Symp. Biol. 1276-85. [PubMed] [Google Scholar]
- 10.Metcalf, W. W., and B. L. Wanner. 1991. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J. Bacteriol. 173587-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarthy, A., S. Michaelis, and J. Beckwith. 1981. Deletion map of the Escherichia coli structural gene for alkaline phosphatase, phoA. J. Bacteriol. 145288-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schweizer, H., and W. Boos. 1983. Cloning of the ugp region containing the structural genes for the pho regulon-dependent sn-glycerol-3-phosphate transport system of Escherichia coli. Mol. Gen. Genet. 192177-186. [DOI] [PubMed] [Google Scholar]
- 13.Schweizer, H., and T. J. Larson. 1987. Cloning and characterization of the aerobic sn-glycerol-3-phosphate dehydrogenase structural gene glpD of Escherichia coli K-12. J. Bacteriol. 169507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torriani, A., and F. Rothman. 1961. Mutants of Escherichia coli constitutive for alkaline phosphatase. J. Bacteriol. 81835-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wanner, B. L. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell Biochem. 5147-54. [DOI] [PubMed] [Google Scholar]
- 16.Wanner, B. L. 1983. Overlapping and separate controls on the phosphate regulon in Escherichia coli K12. J. Mol. Biol. 166283-308. [DOI] [PubMed] [Google Scholar]
- 17.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. I. ASM Press, Washington, DC. [Google Scholar]
- 18.Wanner, B. L., and R. McSharry. 1982. Phosphate-controlled gene expression in Escherichia coli K12 using Mudl-directed lacZ fusions. J. Mol. Biol. 158347-363. [DOI] [PubMed] [Google Scholar]
- 19.White, A. K., and W. W. Metcalf. 2007. Microbial metabolism of reduced phosphorus compounds. Annu. Rev. Microbiol. 61379-400. [DOI] [PubMed] [Google Scholar]
- 20.Yakovleva, G. M., S. K. Kim, and B. L. Wanner. 1998. Phosphate-independent expression of the carbon-phosphorus lyase activity of Escherichia coli. Appl. Microbiol. Biotechnol. 49573-578. [DOI] [PubMed] [Google Scholar]
- 21.Yang, K. C., and W. W. Metcalf. 2004. A new activity for an old enzyme: Escherichia coli bacterial alkaline phosphatase is a phosphite-dependent hydrogenase. Proc. Natl. Acad. Sci. USA 1017919-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]