FIG. 10.
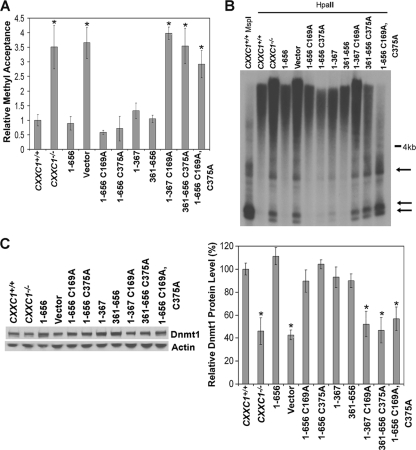
Retention of either the DNA-binding domain or SID of Cfp1 is required to rescue cytosine methylation. (A) Global cytosine methylation levels were determined by methyl acceptance assay for genomic DNA isolated from CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 truncations or mutations (or carrying the empty expression vector). The graph summarizes three independent experiments, and error bars represent standard error. Asterisks denote statistically significant (P < 0.05) differences compared to CXXC1−/− ES cells expressing Cfp11-656. (B) Genomic DNA was isolated from the indicated ES cell lines and digested with MspI or HpaII, and Southern blot analysis was performed using a radiolabeled probe specific for IAP retrovirus repetitive elements. Arrows indicate bands that demonstrate cytosine hypomethylation. (C) Western blot analysis was performed on protein extracts derived from the indicated ES cell lines, using antiserum directed against Dnmt1 (7) and β-actin as a loading control. The graph summarizes data from at least three independent experiments, and error bars represent standard error. The data for the CXXC1+/+, CXXC1−/−, Cfp11-656 (1-656), and vector lanes are reproduced from Fig. 3 to facilitate comparison of rescue activity. Asterisks denote statistically significant (P < 0.05) differences compared to CXXC1−/− ES cells expressing Cfp11-656.