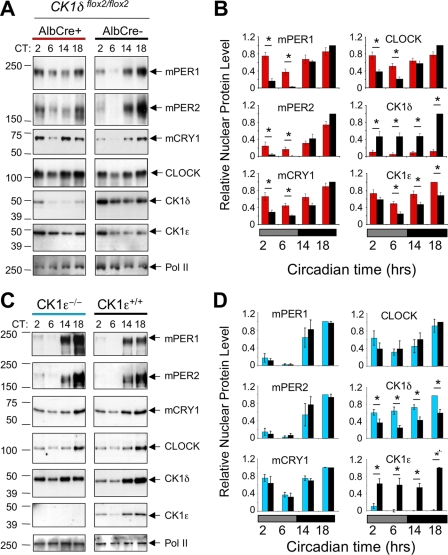
FIG. 5.
Hepatic protein rhythms in CK1-deficient liver. (A) Altered protein rhythms in CK1δ-deficient liver nuclei. Liver nuclei prepared at four time points on the first day in constant darkness were probed to detect circadian proteins by Western blotting. Liver tissue was collected at the indicated circadian times (CT) from CK1δ-deficient livers (AlbCre+; CK1δflox2/flox2) and control livers (AlbCre negative; CK1δflox2/flox2). Rhythms of circadian proteins were present in CK1δ-deficient livers, but the rhythms were blunted in amplitude due to the presence of higher levels of mPER1, mPER2, and CRY1 proteins during the subjective day (CT2 and CT6). The results shown are representative of at least four experiments. (B) Quantitative analysis of hepatic protein rhythms from CK1δ-deficient livers. Relative nuclear protein levels were calculated by densitometric analysis of Western blots; data were first expressed relative to Pol II, and the maximum protein/Pol II ratio in each experiment was set to 1.0. Other values are expressed relative to this maximum. Each panel includes values from four (mCRY1) or five independent experiments generated from different tissue samples. Asterisks indicate significant difference between the genotypes at P < 0.05. (C) Unaltered protein rhythms in liver nuclei from CK1ɛ-deficient mice. Liver nuclei prepared at four circadian times on the first day in constant darkness were probed to detect circadian proteins. Rhythms of circadian proteins were present in CK1ɛ-deficient livers, and these rhythms did not differ from the rhythms in wild-type mice. The results shown are representative of three experiments, each involving different tissue samples. (D) Quantitative analysis of hepatic protein rhythms in CK1ɛ-deficient mice. Nuclear proteins from mice with whole-body disruption of CK1ɛ (CK1ɛ−/−) and wild-type controls (CK1ɛ+/+) were assessed. Three Western blots for each protein (as shown in Fig. 3C) were analyzed as described in panel B, above.